Engineered alterations in RNA editing modulate complex behavior in Drosophila: regulatory diversity of adenosine deaminase acting on RNA (ADAR) targets
- PMID: 21078670
- PMCID: PMC3048717
- DOI: 10.1074/jbc.M110.186817
Engineered alterations in RNA editing modulate complex behavior in Drosophila: regulatory diversity of adenosine deaminase acting on RNA (ADAR) targets
Abstract
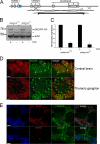
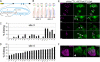
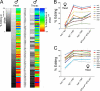
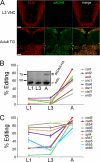
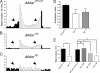
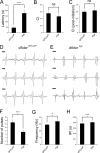
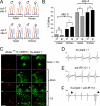
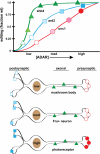
Select proteins involved in electrical and chemical neurotransmission are re-coded at the RNA level via the deamination of particular adenosines to inosine by adenosine deaminases acting on RNA (ADARs). It has been hypothesized that this process, termed RNA editing, acts to "fine-tune" neurophysiological properties in animals and potentially downstream behavioral outputs. However, the extreme phenotypes resulting from deletions of adar loci have precluded investigations into the relationship between ADAR levels, target transcripts, and complex behaviors. Here, we engineer Drosophila hypomorphic for ADAR expression using homologous recombination. A substantial reduction in ADAR activity (>80%) leads to altered circadian motor patterns and abnormal male courtship, although surprisingly, general locomotor coordination is spared. The altered phenotypic landscape in our adar hypomorph is paralleled by an unexpected dichotomous response of ADAR target transcripts, i.e. certain adenosines are minimally affected by dramatic ADAR reduction, whereas editing of others is severely curtailed. Furthermore, we use a novel reporter to map RNA editing activity across the nervous system, and we demonstrate that knockdown of editing in fruitless-expressing neurons is sufficient to modify the male courtship song. Our data demonstrate that network-wide temporal and spatial regulation of ADAR activity can tune the complex system of RNA-editing sites and modulate multiple ethologically relevant behavioral modalities.
Figures
Similar articles
-
Unraveling pleiotropic functions of A-to-I RNA editing in Drosophila.Fly (Austin). 2010 Apr-Jun;4(2):154-8. doi: 10.4161/fly.4.2.11232. Epub 2010 Apr 17. Fly (Austin). 2010. PMID: 20215872
-
Adenosine-to-inosine genetic recoding is required in the adult stage nervous system for coordinated behavior in Drosophila.J Biol Chem. 2009 Nov 6;284(45):31391-400. doi: 10.1074/jbc.M109.035048. Epub 2009 Sep 15. J Biol Chem. 2009. PMID: 19759011 Free PMC article.
-
Zinc Finger RNA-Binding Protein Zn72D Regulates ADAR-Mediated RNA Editing in Neurons.Cell Rep. 2020 May 19;31(7):107654. doi: 10.1016/j.celrep.2020.107654. Cell Rep. 2020. PMID: 32433963 Free PMC article.
-
ADAR RNA editing below the backbone.RNA. 2017 Sep;23(9):1317-1328. doi: 10.1261/rna.060921.117. Epub 2017 May 30. RNA. 2017. PMID: 28559490 Free PMC article. Review.
-
ADAR RNA editing in innate immune response phasing, in circadian clocks and in sleep.Biochim Biophys Acta Gene Regul Mech. 2019 Mar;1862(3):356-369. doi: 10.1016/j.bbagrm.2018.10.011. Epub 2018 Oct 31. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 30391332 Review.
Cited by
-
A-to-I RNA editing: effects on proteins key to neural excitability.Neuron. 2012 May 10;74(3):432-9. doi: 10.1016/j.neuron.2012.04.010. Neuron. 2012. PMID: 22578495 Free PMC article. Review.
-
RNA editing regulates transposon-mediated heterochromatic gene silencing.Nat Commun. 2013;4:2745. doi: 10.1038/ncomms3745. Nat Commun. 2013. PMID: 24201902 Free PMC article.
-
Genome-wide identification and characterization of tissue-specific RNA editing events in D. melanogaster and their potential role in regulating alternative splicing.RNA Biol. 2015;12(12):1391-401. doi: 10.1080/15476286.2015.1107703. RNA Biol. 2015. PMID: 26512413 Free PMC article.
-
ADAR2 enzymes: efficient site-specific RNA editors with gene therapy aspirations.RNA. 2022 Oct;28(10):1281-1297. doi: 10.1261/rna.079266.122. Epub 2022 Jul 21. RNA. 2022. PMID: 35863867 Free PMC article. Review.
-
Linkage of A-to-I RNA Editing in Metazoans and the Impact on Genome Evolution.Mol Biol Evol. 2018 Jan 1;35(1):132-148. doi: 10.1093/molbev/msx274. Mol Biol Evol. 2018. PMID: 29048557 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

