Expression of type II chorionic gonadotropin genes supports a role in the male reproductive system
- PMID: 21078876
- PMCID: PMC3019977
- DOI: 10.1128/MCB.00603-10
Expression of type II chorionic gonadotropin genes supports a role in the male reproductive system
Abstract
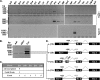
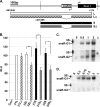
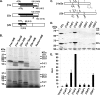
Human chorionic gonadotropin (hCG) is a glycoprotein hormone essential to pregnancy. hCG is heterodimeric and functionally defined by its β subunit. hCGβ evolved from the β subunit of luteinizing hormone in two phases. In the first phase, type I genes (hCGβ3, -5, -7, and -8) acquired changes affecting gene expression and extending the proteins' C terminus. In the second phase, type II genes (hCGβ1 and -2) were formed by the insertion of a DNA element into the type I 5' end. The insertion includes the small noncoding RNA gene snaR-G and has been predicted to drastically change the protein products encoded. We trace the insertion to the common ancestor of the African great apes and show that it contains transcription signals, including snaR-G. Type II transcripts are predominantly expressed in testis. Contrary to predictions, the product of the major mRNA splice form is hCGβ. A novel peptide is encoded by alternatively spliced transcripts. These findings support the view that type II genes evolved in African great apes to function in the male reproductive system.
Figures




Similar articles
-
snaR genes: recent descendants of Alu involved in the evolution of chorionic gonadotropins.Cold Spring Harb Symp Quant Biol. 2009;74:363-73. doi: 10.1101/sqb.2009.74.038. Epub 2009 Dec 22. Cold Spring Harb Symp Quant Biol. 2009. PMID: 20028844 Review.
-
Overexpression of human chorionic gonadotropin causes multiple reproductive defects in transgenic mice.Biol Reprod. 2003 Jul;69(1):338-46. doi: 10.1095/biolreprod.102.013953. Epub 2003 Apr 2. Biol Reprod. 2003. PMID: 12672665
-
A novel two-promoter-one-gene system of the chorionic gonadotropin β gene enables tissue-specific expression.J Mol Endocrinol. 2011 Oct 24;47(3):285-98. doi: 10.1530/JME-11-0026. Print 2011 Dec. J Mol Endocrinol. 2011. PMID: 21821715
-
Luteinizing hormone/chorionic gonadotropin bioactivity in the common marmoset (Callithrix jacchus) is due to a chorionic gonadotropin molecule with a structure intermediate between human chorionic gonadotropin and human luteinizing hormone.Biol Reprod. 1995 Aug;53(2):380-9. doi: 10.1095/biolreprod53.2.380. Biol Reprod. 1995. PMID: 7492691
-
Regulation of chorionic gonadotropin gene expression.Endocr Rev. 1993 Apr;14(2):203-21. doi: 10.1210/edrv-14-2-203. Endocr Rev. 1993. PMID: 8325253 Review. No abstract available.
Cited by
-
The evolution and consequences of snaR family transposition in primates.Mob Genet Elements. 2011 Nov 1;1(4):291-295. doi: 10.4161/mge.18478. Mob Genet Elements. 2011. PMID: 22545241 Free PMC article.
-
Human chorionic gonadotropin beta subunit genes CGB1 and CGB2 are transcriptionally active in ovarian cancer.Int J Mol Sci. 2013 Jun 17;14(6):12650-60. doi: 10.3390/ijms140612650. Int J Mol Sci. 2013. PMID: 23774837 Free PMC article.
-
The Study of the Expression of CGB1 and CGB2 in Human Cancer Tissues.Genes (Basel). 2020 Sep 17;11(9):1082. doi: 10.3390/genes11091082. Genes (Basel). 2020. PMID: 32957442 Free PMC article.
-
The tumor suppressor p53 induces expression of the pregnancy-supporting human chorionic gonadotropin (hCG) CGB7 gene.Cell Cycle. 2011 Nov 1;10(21):3758-67. doi: 10.4161/cc.10.21.17946. Epub 2011 Nov 1. Cell Cycle. 2011. PMID: 22032922 Free PMC article.
References
-
- Adashi, E. Y. 1994. Endocrinology of the ovary. Hum. Reprod. 9:815-827. - PubMed
-
- Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. - PubMed
-
- Berger, P., M. Gruschwitz, G. Spoettl, S. Dirnhofer, S. Madersbacher, R. Gerth, W. E. Merz, E. Plas, and N. Sampson. 2007. Human chorionic gonadotropin (hCG) in the male reproductive tract. Mol. Cell. Endocrinol. 260-262:190-196. - PubMed
-
- Berger, P., W. Kranewitter, S. Madersbacher, R. Gerth, S. Geley, and S. Dirnhofer. 1994. Eutopic production of human chorionic gonadotropin beta (hCG beta) and luteinizing hormone beta (hLH beta) in the human testis. FEBS Lett. 343:229-233. - PubMed
-
- Birken, S., and R. E. Canfield. 1977. Isolation and amino acid sequence of COOH-terminal fragments from the beta subunit of human choriogonadotropin. J. Biol. Chem. 252:5386-5392. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
