CD14 is a coreceptor of Toll-like receptors 7 and 9
- PMID: 21078886
- PMCID: PMC2989773
- DOI: 10.1084/jem.20101111
CD14 is a coreceptor of Toll-like receptors 7 and 9
Abstract
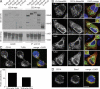
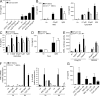
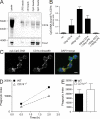
Recognition of pathogens by the innate immune system requires proteins that detect conserved molecular patterns. Nucleic acids are recognized by cytoplasmic sensors as well as by endosomal Toll-like receptors (TLRs). It has become evident that TLRs require additional proteins to be activated by their respective ligands. In this study, we show that CD14 (cluster of differentiation 14) constitutively interacts with the MyD88-dependent TLR7 and TLR9. CD14 was necessary for TLR7- and TLR9-dependent induction of proinflammatory cytokines in vitro and for TLR9-dependent innate immune responses in mice. CD14 associated with TLR9 stimulatory DNA in precipitation experiments and confocal imaging. The absence of CD14 led to reduced nucleic acid uptake in macrophages. Additionally, CD14 played a role in the stimulation of TLRs by viruses. Using various types of vesicular stomatitis virus, we showed that CD14 is dispensable for viral uptake but is required for the triggering of TLR-dependent cytokine responses. These data show that CD14 has a dual role in nucleic acid-mediated TLR activation: it promotes the selective uptake of nucleic acids, and it acts as a coreceptor for endosomal TLR activation.
Figures



References
-
- Andrejeva J., Childs K.S., Young D.F., Carlos T.S., Stock N., Goodbourn S., Randall R.E. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA. 101:17264–17269 10.1073/pnas.0407639101 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

