Annealing helicase 2 (AH2), a DNA-rewinding motor with an HNH motif
- PMID: 21078962
- PMCID: PMC3000258
- DOI: 10.1073/pnas.1011196107
Annealing helicase 2 (AH2), a DNA-rewinding motor with an HNH motif
Abstract
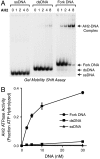
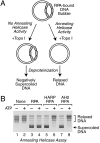
The structure and integrity of DNA is of considerable biological and biomedical importance, and it is therefore critical to identify and to characterize enzymes that alter DNA structure. DNA helicases are ATP-driven motor proteins that unwind DNA. Conversely, HepA-related protein (HARP) protein (also known as SMARCAL1 and DNA-dependent ATPase A) is an annealing helicase that rewinds DNA in an ATP-dependent manner. To date, HARP is the only known annealing helicase. Here we report the identification of a second annealing helicase, which we term AH2, for annealing helicase 2. Like HARP, AH2 catalyzes the ATP-dependent rewinding of replication protein A (RPA)-bound complementary single-stranded DNA, but does not exhibit any detectable helicase activity. Unlike HARP, however, AH2 lacks a conserved RPA-binding domain and does not interact with RPA. In addition, AH2 contains an HNH motif, which is commonly found in bacteria and fungi and is often associated with nuclease activity. AH2 appears to be the only vertebrate protein with an HNH motif. Contrary to expectations, purified AH2 does not exhibit nuclease activity, but it remains possible that AH2 contains a latent nuclease that is activated under specific conditions. These structural and functional differences between AH2 and HARP suggest that different annealing helicases have distinct functions in the cell.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: Mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. - PubMed
-
- Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. - PubMed
-
- Boerkoel CF, et al. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet. 2002;30:215–220. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

