Scalable rule-based modelling of allosteric proteins and biochemical networks
- PMID: 21079669
- PMCID: PMC2973810
- DOI: 10.1371/journal.pcbi.1000975
Scalable rule-based modelling of allosteric proteins and biochemical networks
Abstract
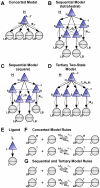
Much of the complexity of biochemical networks comes from the information-processing abilities of allosteric proteins, be they receptors, ion-channels, signalling molecules or transcription factors. An allosteric protein can be uniquely regulated by each combination of input molecules that it binds. This "regulatory complexity" causes a combinatorial increase in the number of parameters required to fit experimental data as the number of protein interactions increases. It therefore challenges the creation, updating, and re-use of biochemical models. Here, we propose a rule-based modelling framework that exploits the intrinsic modularity of protein structure to address regulatory complexity. Rather than treating proteins as "black boxes", we model their hierarchical structure and, as conformational changes, internal dynamics. By modelling the regulation of allosteric proteins through these conformational changes, we often decrease the number of parameters required to fit data, and so reduce over-fitting and improve the predictive power of a model. Our method is thermodynamically grounded, imposes detailed balance, and also includes molecular cross-talk and the background activity of enzymes. We use our Allosteric Network Compiler to examine how allostery can facilitate macromolecular assembly and how competitive ligands can change the observed cooperativity of an allosteric protein. We also develop a parsimonious model of G protein-coupled receptors that explains functional selectivity and can predict the rank order of potency of agonists acting through a receptor. Our methodology should provide a basis for scalable, modular and executable modelling of biochemical networks in systems and synthetic biology.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–52. - PubMed
-
- Tyson JJ, Chen KC, Novak B. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr Opin Cell Biol. 2003;15:221–231. - PubMed
-
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. - PubMed
-
- Bray D. Protein molecules as computational elements in living cells. Nature. 1995;376:307–312. - PubMed
-
- Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–580. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

