Genetic basis of growth adaptation of Escherichia coli after deletion of pgi, a major metabolic gene
- PMID: 21079674
- PMCID: PMC2973815
- DOI: 10.1371/journal.pgen.1001186
Genetic basis of growth adaptation of Escherichia coli after deletion of pgi, a major metabolic gene
Abstract
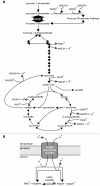
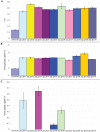
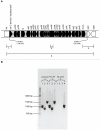
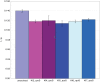
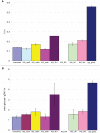
Bacterial survival requires adaptation to different environmental perturbations such as exposure to antibiotics, changes in temperature or oxygen levels, DNA damage, and alternative nutrient sources. During adaptation, bacteria often develop beneficial mutations that confer increased fitness in the new environment. Adaptation to the loss of a major non-essential gene product that cripples growth, however, has not been studied at the whole-genome level. We investigated the ability of Escherichia coli K-12 MG1655 to overcome the loss of phosphoglucose isomerase (pgi) by adaptively evolving ten replicates of E. coli lacking pgi for 50 days in glucose M9 minimal medium and by characterizing endpoint clones through whole-genome re-sequencing and phenotype profiling. We found that 1) the growth rates for all ten endpoint clones increased approximately 3-fold over the 50-day period; 2) two to five mutations arose during adaptation, most frequently in the NADH/NADPH transhydrogenases udhA and pntAB and in the stress-associated sigma factor rpoS; and 3) despite similar growth rates, at least three distinct endpoint phenotypes developed as defined by different rates of acetate and formate secretion. These results demonstrate that E. coli can adapt to the loss of a major metabolic gene product with only a handful of mutations and that adaptation can result in multiple, alternative phenotypes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Herring CD, Raghunathan A, Honisch C, Patel T, Applebee MK, et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 2006;38:1406–1412. - PubMed
-
- Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

