Human-specific evolution and adaptation led to major qualitative differences in the variable receptors of human and chimpanzee natural killer cells
- PMID: 21079681
- PMCID: PMC2973822
- DOI: 10.1371/journal.pgen.1001192
Human-specific evolution and adaptation led to major qualitative differences in the variable receptors of human and chimpanzee natural killer cells
Abstract
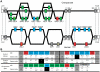
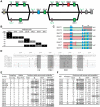
Natural killer (NK) cells serve essential functions in immunity and reproduction. Diversifying these functions within individuals and populations are rapidly-evolving interactions between highly polymorphic major histocompatibility complex (MHC) class I ligands and variable NK cell receptors. Specific to simian primates is the family of Killer cell Immunoglobulin-like Receptors (KIR), which recognize MHC class I and associate with a range of human diseases. Because KIR have considerable species-specificity and are lacking from common animal models, we performed extensive comparison of the systems of KIR and MHC class I interaction in humans and chimpanzees. Although of similar complexity, they differ in genomic organization, gene content, and diversification mechanisms, mainly because of human-specific specialization in the KIR that recognizes the C1 and C2 epitopes of MHC-B and -C. Humans uniquely focused KIR recognition on MHC-C, while losing C1-bearing MHC-B. Reversing this trend, C1-bearing HLA-B46 was recently driven to unprecedented high frequency in Southeast Asia. Chimpanzees have a variety of ancient, avid, and predominantly inhibitory receptors, whereas human receptors are fewer, recently evolved, and combine avid inhibitory receptors with attenuated activating receptors. These differences accompany human-specific evolution of the A and B haplotypes that are under balancing selection and differentially function in defense and reproduction. Our study shows how the qualitative differences that distinguish the human and chimpanzee systems of KIR and MHC class I predominantly derive from adaptations on the human line in response to selective pressures placed on human NK cells by the competing needs of defense and reproduction.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Diefenbach A, Raulet DH. The innate immune response to tumors and its role in the induction of T-cell immunity. Immunol Rev. 2002;188:9–21. - PubMed
-
- Moretta L, Ferlazzo G, Bottino C, Vitale M, Pende D, et al. Effector and regulatory events during natural killer-dendritic cell interactions. Immunol Rev. 2006;214:219–228. - PubMed
-
- Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–510. - PubMed
-
- Moffett-King A. Natural killer cells and pregnancy. Nat Rev Immunol. 2002;2:656–663. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

