C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy
- PMID: 21079745
- PMCID: PMC2975709
- DOI: 10.1371/journal.pone.0015435
C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy
Abstract
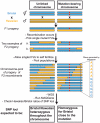
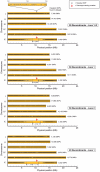
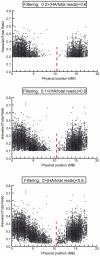
Whole-genome sequencing (WGS) is becoming a fast and cost-effective method to pinpoint molecular lesions in mutagenized genetic model systems, such as Caenorhabditis elegans. As mutagenized strains contain a significant mutational load, it is often still necessary to map mutations to a chromosomal interval to elucidate which of the WGS-identified sequence variants is the phenotype-causing one. We describe here our experience in setting up and testing a simple strategy that incorporates a rapid SNP-based mapping step into the WGS procedure. In this strategy, a mutant retrieved from a genetic screen is crossed with a polymorphic C. elegans strain, individual F2 progeny from this cross is selected for the mutant phenotype, the progeny of these F2 animals are pooled and then whole-genome-sequenced. The density of polymorphic SNP markers is decreased in the region of the phenotype-causing sequence variant and therefore enables its identification in the WGS data. As a proof of principle, we use this strategy to identify the molecular lesion in a mutant strain that produces an excess of dopaminergic neurons. We find that the molecular lesion resides in the Pax-6/Eyeless ortholog vab-3. The strategy described here will further reduce the time between mutant isolation and identification of the molecular lesion.
Conflict of interest statement
Figures

References
-
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

