Nonnative interactions in coupled folding and binding processes of intrinsically disordered proteins
- PMID: 21079758
- PMCID: PMC2973977
- DOI: 10.1371/journal.pone.0015375
Nonnative interactions in coupled folding and binding processes of intrinsically disordered proteins
Abstract
Proteins function by interacting with other molecules, where both native and nonnative interactions play important roles. Native interactions contribute to the stability and specificity of a complex, whereas nonnative interactions mainly perturb the binding kinetics. For intrinsically disordered proteins (IDPs), which do not adopt rigid structures when being free in solution, the role of nonnative interactions may be more prominent in binding processes due to their high flexibilities. In this work, we investigated the effect of nonnative hydrophobic interactions on the coupled folding and binding processes of IDPs and its interplay with chain flexibility by conducting molecular dynamics simulations. Our results showed that the free-energy profiles became rugged, and intermediate states occurred when nonnative hydrophobic interactions were introduced. The binding rate was initially accelerated and subsequently dramatically decreased as the strength of the nonnative hydrophobic interactions increased. Both thermodynamic and kinetic analysis showed that disordered systems were more readily affected by nonnative interactions than ordered systems. Furthermore, it was demonstrated that the kinetic advantage of IDPs ("fly-casting" mechanism) was enhanced by nonnative hydrophobic interactions. The relationship between chain flexibility and protein aggregation is also discussed.
Conflict of interest statement
Figures
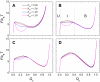
 = 0.29, 0.46, 0.65, and 0.85 by tuning the intramolecular interaction parameter α from 0.1, 1.0, 1.5 to 3.0. The two vertical dashed lines in panel (B) indicate the definition of the unbound state (U), intermediate state (I), and bound state (B). The strength of the nonnative hydrophobic interactions (K
HP) ranges from 0.00 to 1.50.
= 0.29, 0.46, 0.65, and 0.85 by tuning the intramolecular interaction parameter α from 0.1, 1.0, 1.5 to 3.0. The two vertical dashed lines in panel (B) indicate the definition of the unbound state (U), intermediate state (I), and bound state (B). The strength of the nonnative hydrophobic interactions (K
HP) ranges from 0.00 to 1.50.
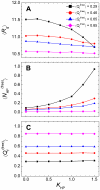
 ), and (C) the average fraction of native contacts (
), and (C) the average fraction of native contacts ( ).
).
 along the binding process when the strength of nonnative hydrophobic interactions was increased: (A–C) K
HP = 0.50, 1.00, and 1.50. (D) Correlation between the average number of nonnative contacts at the intermediate state
along the binding process when the strength of nonnative hydrophobic interactions was increased: (A–C) K
HP = 0.50, 1.00, and 1.50. (D) Correlation between the average number of nonnative contacts at the intermediate state  and
and  . K
HP was set 1.50. (E) Correlation between the average number of nonnative contacts in the bound state
. K
HP was set 1.50. (E) Correlation between the average number of nonnative contacts in the bound state  and
and  . K
HP was set 1.50. (F) Correlation between
. K
HP was set 1.50. (F) Correlation between  and K
HP. The definitions of the intermediate state and bound state are presented in Figure 1.
and K
HP. The definitions of the intermediate state and bound state are presented in Figure 1.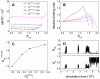
 . (C) Correlation between the K
HP corresponding to the maximum binding rate, K
HP
max-rate, and
. (C) Correlation between the K
HP corresponding to the maximum binding rate, K
HP
max-rate, and  . (D) A typical binding trajectory for the system with
. (D) A typical binding trajectory for the system with  = 0.46 under K
HP = 1.50.
= 0.46 under K
HP = 1.50.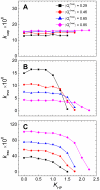
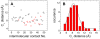

 . Contact distance σhϕ = 7.5 Å was used in (A–C). (D) The sensitivity of the binding rate with respect to nonnative hydrophobic interactions for σhϕ = 5.0 and 7.5 Å.
. Contact distance σhϕ = 7.5 Å was used in (A–C). (D) The sensitivity of the binding rate with respect to nonnative hydrophobic interactions for σhϕ = 5.0 and 7.5 Å.References
-
- Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat Struct Biol. 1997;4:10–19. - PubMed
-
- Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein folding: a synthesis. Proteins. 1995;21:167–195. - PubMed
-
- Ueda Y, Taketomi H, Gō N. Studies on protein folding, unfolding, and fluctuations by computer simulation. II. A. three-dimensional lattice model of lysozyme. Biopolymers. 1978;17:1531–1548.
-
- Gō N. Theoretical studies of protein folding. Annu Rev Biophys Bioeng. 1983;12:183–210. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

