Differential volatile signatures from skin, naevi and melanoma: a novel approach to detect a pathological process
- PMID: 21079799
- PMCID: PMC2973952
- DOI: 10.1371/journal.pone.0013813
Differential volatile signatures from skin, naevi and melanoma: a novel approach to detect a pathological process
Abstract
Background: Early detection of melanoma is of great importance to reduce mortality. Discovering new melanoma biomarkers would improve early detection and diagnosis. Here, we present a novel approach to detect volatile compounds from skin.
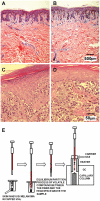
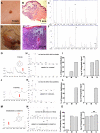
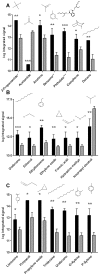
Methods and findings: We used Head Space Solid Phase Micro-Extraction (HS-SPME) and gas chromatography/mass spectrometry (GC/MS) to identify volatile signatures from melanoma, naevi and skin samples. We hypothesized that the metabolic state of tissue alters the profile of volatile compounds. Volatiles released from fresh biopsy tissue of melanoma and benign naevus were compared based on their difference in frequency distribution and their expression level. We also analyzed volatile profiles from frozen tissue, including skin and melanoma.
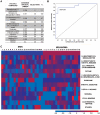
Conclusions: Three volatiles, 4-methyl decane, dodecane and undecane were preferentially expressed in both fresh and frozen melanoma, indicating that they are candidate biomarkers. Twelve candidate biomarkers evaluated by fuzzy logic analysis of frozen samples distinguished melanoma from skin with 89% sensitivity and 90% specificity. Our results demonstrate proof-of-principle that there is differential expression of volatiles in melanoma. Our volatile metabolomic approach will lead to a better understanding of melanoma and can enable development of new diagnostic and treatment strategies based on altered metabolism.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

