Genome-wide comparison of paired fresh frozen and formalin-fixed paraffin-embedded gliomas by custom BAC and oligonucleotide array comparative genomic hybridization: facilitating analysis of archival gliomas
- PMID: 21080181
- PMCID: PMC3059338
- DOI: 10.1007/s00401-010-0773-z
Genome-wide comparison of paired fresh frozen and formalin-fixed paraffin-embedded gliomas by custom BAC and oligonucleotide array comparative genomic hybridization: facilitating analysis of archival gliomas
Abstract
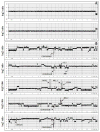
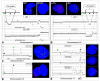
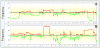
Array comparative genomic hybridization (aCGH) is a powerful tool for detecting DNA copy number alterations (CNA). Because diffuse malignant gliomas are often sampled by small biopsies, formalin-fixed paraffin-embedded (FFPE) blocks are often the only tissue available for genetic analysis; FFPE tissues are also needed to study the intratumoral heterogeneity that characterizes these neoplasms. In this paper, we present a combination of evaluations and technical advances that provide strong support for the ready use of oligonucleotide aCGH on FFPE diffuse gliomas. We first compared aCGH using bacterial artificial chromosome (BAC) arrays in 45 paired frozen and FFPE gliomas, and demonstrate a high concordance rate between FFPE and frozen DNA in an individual clone-level analysis of sensitivity and specificity, assuring that under certain array conditions, frozen and FFPE DNA can perform nearly identically. However, because oligonucleotide arrays offer advantages to BAC arrays in genomic coverage and practical availability, we next developed a method of labeling DNA from FFPE tissue that allows efficient hybridization to oligonucleotide arrays. To demonstrate utility in FFPE tissues, we applied this approach to biphasic anaplastic oligoastrocytomas and demonstrate CNA differences between DNA obtained from the two components. Therefore, BAC and oligonucleotide aCGH can be sensitive and specific tools for detecting CNAs in FFPE DNA, and novel labeling techniques enable the routine use of oligonucleotide arrays for FFPE DNA. In combination, these advances should facilitate genome-wide analysis of rare, small and/or histologically heterogeneous gliomas from FFPE tissues.
Figures



References
-
- Bejjani BA, Shaffer LG. Clinical utility of contemporary molecular cytogenetics. Annu Rev Genomics Hum Genet. 2008;9:71–86. - PubMed
-
- Bussey KJ, Chin K, Lababidi S, Reimers M, Reinhold WC, Kuo WL, Gwadry F, Ajay, Kouros-Mehr H, Fridlyand J, Jain A, Collins C, Nishizuka S, Tonon G, Roschke A, Gehlhaus K, Kirsch I, Scudiero DA, Gray JW, Weinstein JN. Integrating data on DNA copy number with gene expression levels and drug sensitivities in the NCI-60 cell line panel. Mol Cancer Ther. 2006;5:853–867. - PMC - PubMed
-
- Cai WW. Detection of DNA copy-number alterations in complex genomes using array comparative genomic hybridization. Methods Mol Biol. 2007;381:105–119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

