The spliceosomal proteome: at the heart of the largest cellular ribonucleoprotein machine
- PMID: 21080498
- PMCID: PMC3575088
- DOI: 10.1002/pmic.201000354
The spliceosomal proteome: at the heart of the largest cellular ribonucleoprotein machine
Abstract
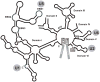
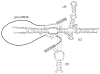
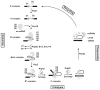
Almost all primary transcripts in higher eukaryotes undergo several splicing events and alternative splicing is a major factor in generating proteomic diversity. Thus, the spliceosome, the ribonucleoprotein assembly that performs splicing, is a highly critical cellular machine and as expected, a very complex one. Indeed, the spliceosome is one of the largest, if not the largest, molecular machine in the cell with over 150 different components in human. A large fraction of the spliceosomal proteome is organized into small nuclear ribonucleoprotein particles by associating with one of the small nuclear RNAs, and the function of many spliceosomal proteins revolve around their association or interaction with the spliceosomal RNAs or the substrate pre-messenger RNAs. In addition to the complex web of protein-RNA interactions, an equally complex network of protein-protein interactions exists in the spliceosome, which includes a number of large, conserved proteins with critical functions in the spliceosomal catalytic core. These include the largest conserved nuclear protein, Prp8, which plays a critical role in spliceosomal function in a hitherto unknown manner. Taken together, the large spliceosomal proteome and its dynamic nature has made it a highly challenging system to study, and at the same time, provides an exciting example of the evolution of a proteome around a backbone of primordial RNAs likely dating from the RNA World.
Copyright © 2010 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
The author declares no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

