Mining the bitter melon (momordica charantia l.) seed transcriptome by 454 analysis of non-normalized and normalized cDNA populations for conjugated fatty acid metabolism-related genes
- PMID: 21080948
- PMCID: PMC3012625
- DOI: 10.1186/1471-2229-10-250
Mining the bitter melon (momordica charantia l.) seed transcriptome by 454 analysis of non-normalized and normalized cDNA populations for conjugated fatty acid metabolism-related genes
Abstract
Background: Seeds of Momordica charantia (bitter melon) produce high levels of eleostearic acid, an unusual conjugated fatty acid with industrial value. Deep sequencing of non-normalized and normalized cDNAs from developing bitter melon seeds was conducted to uncover key genes required for biotechnological transfer of conjugated fatty acid production to existing oilseed crops. It is expected that these studies will also provide basic information regarding the metabolism of other high-value novel fatty acids.
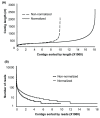
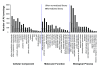
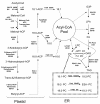
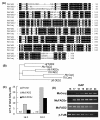
Results: Deep sequencing using 454 technology with non-normalized and normalized cDNA libraries prepared from bitter melon seeds at 18 DAP resulted in the identification of transcripts for the vast majority of known genes involved in fatty acid and triacylglycerol biosynthesis. The non-normalized library provided a transcriptome profile of the early stage in seed development that highlighted the abundance of transcripts for genes encoding seed storage proteins as well as for a number of genes for lipid metabolism-associated polypeptides, including Δ12 oleic acid desaturases and fatty acid conjugases, class 3 lipases, acyl-carrier protein, and acyl-CoA binding protein. Normalization of cDNA by use of a duplex-specific nuclease method not only increased the overall discovery of genes from developing bitter melon seeds, but also resulted in the identification of 345 contigs with homology to 189 known lipid genes in Arabidopsis. These included candidate genes for eleostearic acid metabolism such as diacylglycerol acyltransferase 1 and 2, and a phospholipid:diacylglycerol acyltransferase 1-related enzyme. Transcripts were also identified for a novel FAD2 gene encoding a functional Δ12 oleic acid desaturase with potential implications for eleostearic acid biosynthesis.
Conclusions: 454 deep sequencing, particularly with normalized cDNA populations, was an effective method for mining of genes associated with eleostearic acid metabolism in developing bitter melon seeds. The transcriptomic data presented provide a resource for the study of novel fatty acid metabolism and for the biotechnological production of conjugated fatty acids and possibly other novel fatty acids in established oilseed crops.
Figures


References
-
- Sonntag NOV. In: Bailey's Industrial Oil and Fat Products. Swern D, editor. New York: John Wiley & Sons; 1979. Composition and characteristics of individual fats and oils; pp. 289–477.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

