Analyses of genome architecture and gene expression reveal novel candidate virulence factors in the secretome of Phytophthora infestans
- PMID: 21080964
- PMCID: PMC3091767
- DOI: 10.1186/1471-2164-11-637
Analyses of genome architecture and gene expression reveal novel candidate virulence factors in the secretome of Phytophthora infestans
Abstract
Background: Phytophthora infestans is the most devastating pathogen of potato and a model organism for the oomycetes. It exhibits high evolutionary potential and rapidly adapts to host plants. The P. infestans genome experienced a repeat-driven expansion relative to the genomes of Phytophthora sojae and Phytophthora ramorum and shows a discontinuous distribution of gene density. Effector genes, such as members of the RXLR and Crinkler (CRN) families, localize to expanded, repeat-rich and gene-sparse regions of the genome. This distinct genomic environment is thought to contribute to genome plasticity and host adaptation.
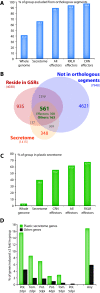
Results: We used in silico approaches to predict and describe the repertoire of P. infestans secreted proteins (the secretome). We defined the "plastic secretome" as a subset of the genome that (i) encodes predicted secreted proteins, (ii) is excluded from genome segments orthologous to the P. sojae and P. ramorum genomes and (iii) is encoded by genes residing in gene sparse regions of P. infestans genome. Although including only ~3% of P. infestans genes, the plastic secretome contains ~62% of known effector genes and shows >2 fold enrichment in genes induced in planta. We highlight 19 plastic secretome genes induced in planta but distinct from previously described effectors. This list includes a trypsin-like serine protease, secreted oxidoreductases, small cysteine-rich proteins and repeat containing proteins that we propose to be novel candidate virulence factors.
Conclusions: This work revealed a remarkably diverse plastic secretome. It illustrates the value of combining genome architecture with comparative genomics to identify novel candidate virulence factors from pathogen genomes.
Figures






References
-
- Kirk WW, Abu-El Samen F, Tumbalam P, Wharton P, Douches D, Thill CA, Thompson A. Impact of Different US Genotypes of Phytophthora infestans on Potato Seed Tuber Rot and Plant Emergence in a Range of Cultivars and Advanced Breeding Lines. Potato Research. 2009;52:121–140. doi: 10.1007/s11540-009-9125-6. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

