Integrated genomics of susceptibility to alkylator-induced leukemia in mice
- PMID: 21080971
- PMCID: PMC3018144
- DOI: 10.1186/1471-2164-11-638
Integrated genomics of susceptibility to alkylator-induced leukemia in mice
Abstract
Background: Therapy-related acute myeloid leukemia (t-AML) is a secondary, generally incurable, malignancy attributable to chemotherapy exposure. Although there is a genetic component to t-AML susceptibility in mice, the relevant loci and the mechanism(s) by which they contribute to t-AML are largely unknown. An improved understanding of susceptibility factors and the biological processes in which they act may lead to the development of t-AML prevention strategies.
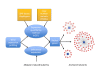
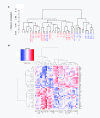
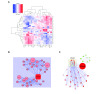
Results: In this work we applied an integrated genomics strategy in inbred strains of mice to find novel factors that might contribute to susceptibility. We found that the pre-exposure transcriptional state of hematopoietic stem/progenitor cells predicts susceptibility status. More than 900 genes were differentially expressed between susceptible and resistant strains and were highly enriched in the apoptotic program, but it remained unclear which genes, if any, contribute directly to t-AML susceptibility. To address this issue, we integrated gene expression data with genetic information, including single nucleotide polymorphisms (SNPs) and DNA copy number variants (CNVs), to identify genetic networks underlying t-AML susceptibility. The 30 t-AML susceptibility networks we found are robust: they were validated in independent, previously published expression data, and different analytical methods converge on them. Further, the networks are enriched in genes involved in cell cycle and DNA repair (pathways not discovered in traditional differential expression analysis), suggesting that these processes contribute to t-AML susceptibility. Within these networks, the putative regulators (e.g., Parp2, Casp9, Polr1b) are the most likely to have a non-redundant role in the pathogenesis of t-AML. While identifying these networks, we found that current CNVR and SNP-based haplotype maps in mice represented distinct sources of genetic variation contributing to expression variation, implying that mapping studies utilizing either source alone will have reduced sensitivity.
Conclusion: The identification and prioritization of genes and networks not previously implicated in t-AML generates novel hypotheses on the biology and treatment of this disease that will be the focus of future research.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

