A stochastic model for chemotaxis based on the ordered extension of pseudopods
- PMID: 21081083
- PMCID: PMC2980707
- DOI: 10.1016/j.bpj.2010.09.042
A stochastic model for chemotaxis based on the ordered extension of pseudopods
Abstract
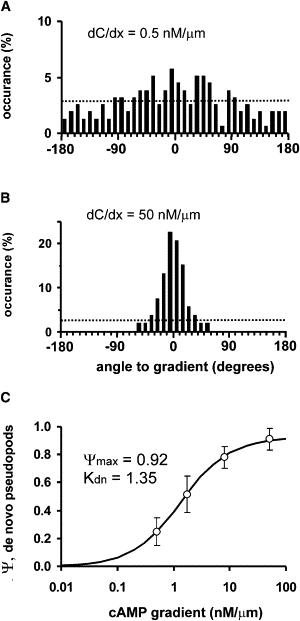
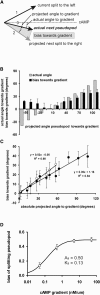
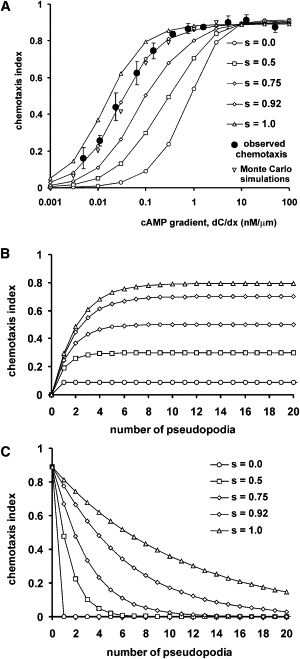
Many amoeboid cells move by extending pseudopods. Here I present a new stochastic model for chemotaxis that is based on pseudopod extensions by Dictyostelium cells. In the absence of external cues, pseudopod extension is highly ordered with two types of pseudopods: de novo formation of a pseudopod at the cell body in random directions, and alternating right/left splitting of an existing pseudopod that leads to a persistent zig-zag trajectory. We measured the directional probabilities of the extension of splitting and de novo pseudopods in chemoattractant gradients with different steepness. Very shallow cAMP gradients can bias the direction of splitting pseudopods, but the bias is not perfect. Orientation of de novo pseudopods require much steeper cAMP gradients and can be more precise. These measured probabilities of pseudopod directions were used to obtain an analytical model for chemotaxis of cell populations. Measured chemotaxis of wild-type cells and mutants with specific defects in these stochastic pseudopod properties are similar to predictions of the model. These results show that combining splitting and de novo pseudopods is a very effective way for cells to obtain very high sensitivity to stable gradient and still be responsive to changes in the direction of the gradient.
Copyright © 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

