Multiple neurofilament subunits are present in lamprey CNS
- PMID: 21081119
- PMCID: PMC3018571
- DOI: 10.1016/j.brainres.2010.11.037
Multiple neurofilament subunits are present in lamprey CNS
Abstract
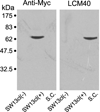
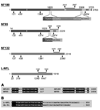
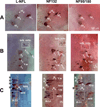
In mammals, there are three neurofilament (NF) subunits (NF-L, NF-M, and NF-H), but it was thought that only a single NF, NF180, exists in lamprey. However, NF180 lacked the ability to self-assemble, suggesting that like mammalian NFs, lamprey NFs are heteropolymers, and that additional NF subunits may exist. The present study provides evidence for the existence of a lamprey NF-L homolog (L-NFL). Genes encoding two new NF-M isoforms (NF132 and NF95) also have been isolated and characterized. With NF180, this makes three NF-M-like isoforms. In situ hybridization showed that all three newly cloned NFs are expressed in spinal cord neurons and in spinal-projecting neurons of the brainstem. Like NF180, there were no KSP multiphosphorylation repeat motifs in the tail regions of NF132 or NF95. NF95 was highly identical to homologous parts of NF180, sharing 2 common pieces of DNA with it. Northern blots suggested that NF95 may be expressed at very low levels in older larvae. The presence of L-NFL in lamprey CNS may support the hypothesis that as in mammals, NFs in lamprey are obligate heteropolymers, in which NF-L is a required subunit.
Copyright © 2010 Elsevier B.V. All rights reserved.
Figures










Similar articles
-
Assembly properties of lamprey neurofilament subunits and their expression after spinal cord transection.J Comp Neurol. 2011 Dec 15;519(18):3657-71. doi: 10.1002/cne.22673. J Comp Neurol. 2011. PMID: 21618230 Free PMC article.
-
Lamprey neurofilaments combine in one subunit the features of each mammalian NF triplet protein but are highly phosphorylated only in large axons.J Neurosci. 1989 Feb;9(2):698-709. doi: 10.1523/JNEUROSCI.09-02-00698.1989. J Neurosci. 1989. PMID: 2493079 Free PMC article.
-
The single neurofilament subunit of lamprey may need another element for filament assembly.J Comp Neurol. 2004 Mar 29;471(2):188-200. doi: 10.1002/cne.20026. J Comp Neurol. 2004. PMID: 14986312
-
Contributions of identifiable neurons and neuron classes to lamprey vertebrate neurobiology.Prog Neurobiol. 2001 Mar;63(4):441-66. doi: 10.1016/s0301-0082(00)00050-2. Prog Neurobiol. 2001. PMID: 11163686 Review.
-
Neurofilaments and Neurofilament Proteins in Health and Disease.Cold Spring Harb Perspect Biol. 2017 Apr 3;9(4):a018309. doi: 10.1101/cshperspect.a018309. Cold Spring Harb Perspect Biol. 2017. PMID: 28373358 Free PMC article. Review.
Cited by
-
Activated Erk Is an Early Retrograde Signal After Spinal Cord Injury in the Lamprey.Front Neurosci. 2020 Nov 5;14:580692. doi: 10.3389/fnins.2020.580692. eCollection 2020. Front Neurosci. 2020. PMID: 33250705 Free PMC article.
-
Neurogenesis in the lamprey central nervous system following spinal cord transection.J Comp Neurol. 2014 Apr 15;522(6):1316-32. doi: 10.1002/cne.23485. J Comp Neurol. 2014. PMID: 24151158 Free PMC article.
-
Protein synthetic machinery and mRNA in regenerating tips of spinal cord axons in lamprey.J Comp Neurol. 2016 Dec 1;524(17):3614-3640. doi: 10.1002/cne.24020. Epub 2016 May 19. J Comp Neurol. 2016. PMID: 27120118 Free PMC article.
-
Assembly properties of lamprey neurofilament subunits and their expression after spinal cord transection.J Comp Neurol. 2011 Dec 15;519(18):3657-71. doi: 10.1002/cne.22673. J Comp Neurol. 2011. PMID: 21618230 Free PMC article.
-
Antisense Morpholino Oligonucleotides Reduce Neurofilament Synthesis and Inhibit Axon Regeneration in Lamprey Reticulospinal Neurons.PLoS One. 2015 Sep 14;10(9):e0137670. doi: 10.1371/journal.pone.0137670. eCollection 2015. PLoS One. 2015. PMID: 26366578 Free PMC article.
References
-
- Asch WS, Leake D, Canger AK, Passini MA, Argenton F, Schechter N. Cloning of zebrafish neurofilament cDNAs for plasticin and gefiltin: increased mRNA expression in ganglion cells after optic nerve injury. J Neurochem. 1998;71:20–32. - PubMed
-
- Bennett GS. Changes in intermediate filament composition during neurogenesis. Curr Top Dev Biol. 1987;21:151–183. - PubMed
-
- Breathnach R, Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. - PubMed
-
- Bullock TH, Moore JK, Fields RD. Evolution of myelin sheaths: both lamprey and hagfish lack myelin. Neurosci Lett. 1984;48:145–148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

