Taming the complexity of protein folding
- PMID: 21081274
- PMCID: PMC3042729
- DOI: 10.1016/j.sbi.2010.10.006
Taming the complexity of protein folding
Abstract
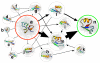
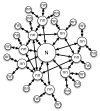
Protein folding is an important problem in structural biology with significant medical implications, particularly for misfolding disorders like Alzheimer's disease. Solving the folding problem will ultimately require a combination of theory and experiment, with theoretical models providing a comprehensive view of folding and experiments grounding these models in reality. Here we review progress towards this goal over the past decade, with an emphasis on recent theoretical advances that are empowering chemically detailed models of folding and the new results these technologies are providing. In particular, we discuss new insights made possible by Markov state models (MSMs), including the role of non-native contacts and the hub-like character of protein folded states.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures

References
-
- Uversky VN. Intrinsic disorder in proteins associated with neurodegenerative diseases. Front Biosci. 2009;14:5188–5238. - PubMed
-
- Schütte C, Fischer A, Huisinga W, Deuflhard P. A direct approach to conformational dynamics based on hybrid Monte Carlo. J Comput Phys. 1999;151:146–168.
-
- Noe F, Fischer S. Transition networks for modeling the kinetics of conformational change in macromolecules. Curr Opin Struct Biol. 2008;18:154–162. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

