ERAD components in organisms with complex red plastids suggest recruitment of a preexisting protein transport pathway for the periplastid membrane
- PMID: 21081314
- PMCID: PMC3045029
- DOI: 10.1093/gbe/evq074
ERAD components in organisms with complex red plastids suggest recruitment of a preexisting protein transport pathway for the periplastid membrane
Abstract
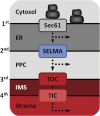
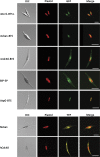
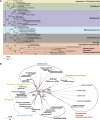
The plastids of cryptophytes, haptophytes, and heterokontophytes (stramenopiles) (together once known as chromists) are surrounded by four membranes, reflecting the origin of these plastids through secondary endosymbiosis. They share this trait with apicomplexans, which are alveolates, the plastids of which have been suggested to stem from the same secondary symbiotic event and therefore form a phylogenetic clade, the chromalveolates. The chromists are quantitatively the most important eukaryotic contributors to primary production in marine ecosystems. The mechanisms of protein import across their four plastid membranes are still poorly understood. Components of an endoplasmic reticulum-associated degradation (ERAD) machinery in cryptophytes, partially encoded by the reduced genome of the secondary symbiont (the nucleomorph), are implicated in protein transport across the second outermost plastid membrane. Here, we show that the haptophyte Emiliania huxleyi, like cryptophytes, stramenopiles, and apicomplexans, possesses a nuclear-encoded symbiont-specific ERAD machinery (SELMA, symbiont-specific ERAD-like machinery) in addition to the host ERAD system, with targeting signals that are able to direct green fluorescent protein or yellow fluorescent protein to the predicted cellular localization in transformed cells of the stramenopile Phaeodactylum tricornutum. Phylogenies of the duplicated ERAD factors reveal that all SELMA components trace back to a red algal origin. In contrast, the host copies of cryptophytes and haptophytes associate with the green lineage to the exclusion of stramenopiles and alveolates. Although all chromalveolates with four membrane-bound plastids possess the SELMA system, this has apparently not arisen in a single endosymbiotic event. Thus, our data do not support the chromalveolate hypothesis.
Figures

Similar articles
-
Three old and one new: protein import into red algal-derived plastids surrounded by four membranes.Protoplasma. 2013 Oct;250(5):1013-23. doi: 10.1007/s00709-013-0498-7. Epub 2013 Apr 24. Protoplasma. 2013. PMID: 23612938 Review.
-
Distribution of the SELMA translocon in secondary plastids of red algal origin and predicted uncoupling of ubiquitin-dependent translocation from degradation.Eukaryot Cell. 2012 Dec;11(12):1472-81. doi: 10.1128/EC.00183-12. Epub 2012 Oct 5. Eukaryot Cell. 2012. PMID: 23042132 Free PMC article.
-
Der1-mediated preprotein import into the periplastid compartment of chromalveolates?Mol Biol Evol. 2007 Apr;24(4):918-28. doi: 10.1093/molbev/msm008. Epub 2007 Jan 22. Mol Biol Evol. 2007. PMID: 17244602
-
Chromera velia, endosymbioses and the rhodoplex hypothesis--plastid evolution in cryptophytes, alveolates, stramenopiles, and haptophytes (CASH lineages).Genome Biol Evol. 2014 Mar;6(3):666-84. doi: 10.1093/gbe/evu043. Genome Biol Evol. 2014. PMID: 24572015 Free PMC article.
-
New Insights Into Roles of Ubiquitin Modification in Regulating Plastids and Other Endosymbiotic Organelles.Int Rev Cell Mol Biol. 2016;325:1-33. doi: 10.1016/bs.ircmb.2016.02.007. Epub 2016 Apr 19. Int Rev Cell Mol Biol. 2016. PMID: 27241217 Review.
Cited by
-
Three old and one new: protein import into red algal-derived plastids surrounded by four membranes.Protoplasma. 2013 Oct;250(5):1013-23. doi: 10.1007/s00709-013-0498-7. Epub 2013 Apr 24. Protoplasma. 2013. PMID: 23612938 Review.
-
Metabolic Innovations Underpinning the Origin and Diversification of the Diatom Chloroplast.Biomolecules. 2019 Jul 30;9(8):322. doi: 10.3390/biom9080322. Biomolecules. 2019. PMID: 31366180 Free PMC article. Review.
-
A Plastid Protein That Evolved from Ubiquitin and Is Required for Apicoplast Protein Import in Toxoplasma gondii.mBio. 2017 Jun 27;8(3):e00950-17. doi: 10.1128/mBio.00950-17. mBio. 2017. PMID: 28655825 Free PMC article.
-
Shedding light on the expansion and diversification of the Cdc48 protein family during the rise of the eukaryotic cell.BMC Evol Biol. 2016 Oct 18;16(1):215. doi: 10.1186/s12862-016-0790-1. BMC Evol Biol. 2016. PMID: 27756227 Free PMC article.
-
Distribution of the SELMA translocon in secondary plastids of red algal origin and predicted uncoupling of ubiquitin-dependent translocation from degradation.Eukaryot Cell. 2012 Dec;11(12):1472-81. doi: 10.1128/EC.00183-12. Epub 2012 Oct 5. Eukaryot Cell. 2012. PMID: 23042132 Free PMC article.
References
-
- Apt KE, Kroth-Pancic PG, Grossman AR. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol Gen Genet. 1996;252:572–579. - PubMed
-
- Armbrust EV, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. - PubMed
-
- Baurain D, et al. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol Biol Evol. 2010;27:1698–1709. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

