ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function
- PMID: 21081504
- PMCID: PMC3024787
- DOI: 10.1074/jbc.M110.171975
ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function
Abstract
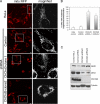
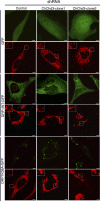
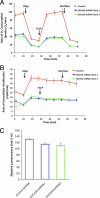
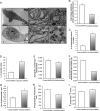
The mitochondrial inner membrane (IM) serves as the site for ATP production by hosting the oxidative phosphorylation complex machinery most notably on the crista membranes. Disruption of the crista structure has been implicated in a variety of cardiovascular and neurodegenerative diseases. Here, we characterize ChChd3, a previously identified PKA substrate of unknown function (Schauble, S., King, C. C., Darshi, M., Koller, A., Shah, K., and Taylor, S. S. (2007) J. Biol. Chem. 282, 14952-14959), and show that it is essential for maintaining crista integrity and mitochondrial function. In the mitochondria, ChChd3 is a peripheral protein of the IM facing the intermembrane space. RNAi knockdown of ChChd3 in HeLa cells resulted in fragmented mitochondria, reduced OPA1 protein levels and impaired fusion, and clustering of the mitochondria around the nucleus along with reduced growth rate. Both the oxygen consumption and glycolytic rates were severely restricted. Ultrastructural analysis of these cells revealed aberrant mitochondrial IM structures with fragmented and tubular cristae or loss of cristae, and reduced crista membrane. Additionally, the crista junction opening diameter was reduced to 50% suggesting remodeling of cristae in the absence of ChChd3. Analysis of the ChChd3-binding proteins revealed that ChChd3 interacts with the IM proteins mitofilin and OPA1, which regulate crista morphology, and the outer membrane protein Sam50, which regulates import and assembly of β-barrel proteins on the outer membrane. Knockdown of ChChd3 led to almost complete loss of both mitofilin and Sam50 proteins and alterations in several mitochondrial proteins, suggesting that ChChd3 is a scaffolding protein that stabilizes protein complexes involved in maintaining crista architecture and protein import and is thus essential for maintaining mitochondrial structure and function.
Figures





Similar articles
-
Targeting and import mechanism of coiled-coil helix coiled-coil helix domain-containing protein 3 (ChChd3) into the mitochondrial intermembrane space.J Biol Chem. 2012 Nov 16;287(47):39480-91. doi: 10.1074/jbc.M112.387696. Epub 2012 Sep 27. J Biol Chem. 2012. PMID: 23019327 Free PMC article.
-
The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11.FEBS Lett. 2007 Jul 24;581(18):3545-9. doi: 10.1016/j.febslet.2007.06.052. Epub 2007 Jun 27. FEBS Lett. 2007. PMID: 17624330
-
MICOS assembly controls mitochondrial inner membrane remodeling and crista junction redistribution to mediate cristae formation.EMBO J. 2020 Jul 15;39(14):e104105. doi: 10.15252/embj.2019104105. Epub 2020 Jun 22. EMBO J. 2020. PMID: 32567732 Free PMC article.
-
Mitofilin complexes: conserved organizers of mitochondrial membrane architecture.Biol Chem. 2012 Nov;393(11):1247-61. doi: 10.1515/hsz-2012-0239. Biol Chem. 2012. PMID: 23109542 Review.
-
Cardiolipin and mitochondrial cristae organization.Biochim Biophys Acta Biomembr. 2017 Jun;1859(6):1156-1163. doi: 10.1016/j.bbamem.2017.03.013. Epub 2017 Mar 20. Biochim Biophys Acta Biomembr. 2017. PMID: 28336315 Free PMC article. Review.
Cited by
-
Bcl-xL-mediated remodeling of rod and cone synaptic mitochondria after postnatal lead exposure: electron microscopy, tomography and oxygen consumption.Mol Vis. 2012;18:3029-48. Epub 2012 Dec 20. Mol Vis. 2012. PMID: 23288995 Free PMC article.
-
Loss of functional OPA1 unbalances redox state: implications in dominant optic atrophy pathogenesis.Ann Clin Transl Neurol. 2016 Apr 25;3(6):408-21. doi: 10.1002/acn3.305. eCollection 2016 Jun. Ann Clin Transl Neurol. 2016. PMID: 27547769 Free PMC article.
-
The connection between inner membrane topology and mitochondrial function.J Mol Cell Cardiol. 2013 Sep;62:51-7. doi: 10.1016/j.yjmcc.2013.05.001. Epub 2013 May 12. J Mol Cell Cardiol. 2013. PMID: 23672826 Free PMC article. Review.
-
Heart failure and mitochondrial dysfunction: the role of mitochondrial fission/fusion abnormalities and new therapeutic strategies.J Cardiovasc Pharmacol. 2014 Mar;63(3):196-206. doi: 10.1097/01.fjc.0000432861.55968.a6. J Cardiovasc Pharmacol. 2014. PMID: 23884159 Free PMC article. Review.
-
Targeting and import mechanism of coiled-coil helix coiled-coil helix domain-containing protein 3 (ChChd3) into the mitochondrial intermembrane space.J Biol Chem. 2012 Nov 16;287(47):39480-91. doi: 10.1074/jbc.M112.387696. Epub 2012 Sep 27. J Biol Chem. 2012. PMID: 23019327 Free PMC article.
References
-
- Chan D. C. (2006) Annu. Rev. Cell Dev. Biol. 22, 79–99 - PubMed
-
- Hoppins S., Lackner L., Nunnari J. (2007) Annu. Rev. Biochem. 76, 751–780 - PubMed
-
- Olichon A., Emorine L. J., Descoins E., Pelloquin L., Brichese L., Gas N., Guillou E., Delettre C., Valette A., Hamel C. P., Ducommun B., Lenaers G., Belenguer P. (2002) FEBS Lett. 523, 171–176 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

