Unraveling the evolution of auxin signaling
- PMID: 21081694
- PMCID: PMC3075796
- DOI: 10.1104/pp.110.168161
Unraveling the evolution of auxin signaling
Abstract
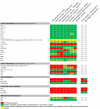
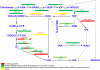
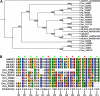
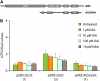
Auxin signaling is central to plant growth and development, yet hardly anything is known about its evolutionary origin. While the presence of key players in auxin signaling has been analyzed in various land plant species, similar analyses in the green algal lineages are lacking. Here, we survey the key players in auxin biology in the available genomes of Chlorophyta species. We found that the genetic potential for auxin biosynthesis and AUXIN1 (AUX1)/LIKE AUX1- and P-GLYCOPROTEIN/ATP-BINDING CASSETTE subfamily B-dependent transport is already present in several single-celled and colony-forming Chlorophyta species. In addition, our analysis of expressed sequence tag libraries from Coleochaete orbicularis and Spirogyra pratensis, green algae of the Streptophyta clade that are evolutionarily closer to the land plants than those of the Chlorophyta clade, revealed the presence of partial AUXIN RESPONSE FACTORs and/or AUXIN/INDOLE-3-ACETIC ACID proteins (the key factors in auxin signaling) and PIN-FORMED-like proteins (the best-characterized auxin-efflux carriers). While the identification of these possible AUXIN RESPONSE FACTOR- and AUXIN/INDOLE-3-ACETIC ACID precursors and putative PIN-FORMED orthologs calls for a deeper investigation of their evolution after sequencing more intermediate genomes, it emphasizes that the canonical auxin response machinery and auxin transport mechanisms were, at least in part, already present before plants "moved" to land habitats.
Figures




References
-
- Beer LL, Boyd ES, Peters JW, Posewitz MC. (2009) Engineering algae for biohydrogen and biofuel production. Curr Opin Biotechnol 20: 264–271 - PubMed
-
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 - PubMed
-
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

