Trisomy-21 gene dosage over-expression of miRNAs results in the haploinsufficiency of specific target proteins
- PMID: 21081842
- PMCID: PMC3073250
- DOI: 10.4161/rna.7.5.12685
Trisomy-21 gene dosage over-expression of miRNAs results in the haploinsufficiency of specific target proteins
Abstract
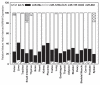
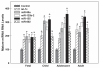
Down syndrome (DS) or Trisomy 21 (Ts21) is caused by the presence of an extra copy of all or part of human chromosome 21 (Hsa21) and is the most frequent survivable congenital chromosomal abnormality. Bioinformatic annotation has established that Hsa21 harbors more than 400 genes, including five microRNA (miRNA) genes (miR-99a, let-7c, miR-125b-2, miR-155, and miR-802). MiRNAs are endogenous, single-stranded, small non-coding RNA molecules that regulate gene expression by interacting with specific recognition elements harbored within the 3'-untranslated region (3'-UTR) of mRNAs and subsequently target these mRNAs for translational repression or destabilization. MiRNA expression profiling, miRNA RT-PCR, and miRNA in situ hybridization experiments have demonstrated that Hsa21-derived miRNAs were over-expressed in fetal brain and heart specimens isolated from individuals with DS. We now propose that Ts21 gene dosage over-expression of Hsa21-derived miRNAs in DS individuals result in the decreased expression of specific target proteins (i.e. haploinsufficiency) that contribute, in part, to the DS phenotype.
Figures
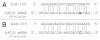
References
-
- LeJeune J, Gautier M, Turpin R. Study of somatic chromosomes from mongoloid children. Comptes Renus de l'Academic les Sciences. 1959;248:1721–1722. - PubMed
-
- Muller F, Rebiffe M, Taillandier A, Oury JF, Mornet E. Parental origin of the extra chromosome in prenatally diagnosed fetal trisomy 21. Hum Genet. 2000;106:340–344. - PubMed
-
- Hassold T, Hunt P. To err (meiotically) is human: The genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. - PubMed
-
- Canfield MA, Honein MA, Yuskiv N, Xing J, Mai CT, Collins JS, et al. National estimates and race/ethnic-specific variation of selected birth defects in the United States 1999–2001. Birth Defects Res A Clin Mol Teratol. 2006;76:747–756. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
