Generation of humoral immune responses to multi-allele PfAMA1 vaccines; effect of adjuvant and number of component alleles on the breadth of response
- PMID: 21082025
- PMCID: PMC2972715
- DOI: 10.1371/journal.pone.0015391
Generation of humoral immune responses to multi-allele PfAMA1 vaccines; effect of adjuvant and number of component alleles on the breadth of response
Abstract
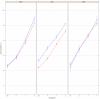
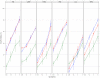
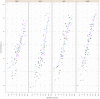
There is increasing interest in multi-allele vaccines to overcome strain-specificity against polymorphic vaccine targets such as Apical Membrane Antigen 1 (AMA1). These have been shown to induce broad inhibitory antibodies in vitro and formed the basis for the design of three Diversity-Covering (DiCo) proteins with similar immunological effects. The antibodies produced are to epitopes that are shared between vaccine alleles and theoretically, increasing the number of component AMA1 alleles is expected to broaden the antibody response. A plateau effect could however impose a limit on the number of alleles needed to achieve the broadest specificity. Moreover, production cost and the vaccine formulation process would limit the number of component alleles. In this paper, we compare rabbit antibody responses elicited with multi-allele vaccines incorporating seven (three DiCos and four natural AMA1 alleles) and three (DiCo mix) antigens for gains in broadened specificity. We also investigate the effect of three adjuvant platforms on antigen specificity and antibody functionality. Our data confirms a broadened response after immunisation with DiCo mix in all three adjuvants. Higher antibody titres were elicited with either CoVaccine HT™ or Montanide ISA 51, resulting in similar in vitro inhibition (65-82%) of five out of six culture-adapted P. falciparum strains. The antigen binding specificities of elicited antibodies were also similar and independent of the adjuvant used or the number of vaccine component alleles. Thus neither the four extra antigens nor adjuvant had any observable benefits with respect to specificity broadening, although adjuvant choice influenced the absolute antibody levels and thus the extent of parasite inhibition. Our data confirms the feasibility and potential of multi-allele PfAMA1 formulations, and highlights the need for adjuvants with improved antibody potentiation properties for AMA1-based vaccines.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

