Integration of deep transcriptome and proteome analyses reveals the components of alkaloid metabolism in opium poppy cell cultures
- PMID: 21083930
- PMCID: PMC3095332
- DOI: 10.1186/1471-2229-10-252
Integration of deep transcriptome and proteome analyses reveals the components of alkaloid metabolism in opium poppy cell cultures
Abstract
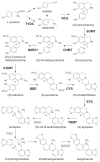
Background: Papaver somniferum (opium poppy) is the source for several pharmaceutical benzylisoquinoline alkaloids including morphine, the codeine and sanguinarine. In response to treatment with a fungal elicitor, the biosynthesis and accumulation of sanguinarine is induced along with other plant defense responses in opium poppy cell cultures. The transcriptional induction of alkaloid metabolism in cultured cells provides an opportunity to identify components of this process via the integration of deep transcriptome and proteome databases generated using next-generation technologies.
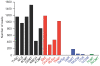
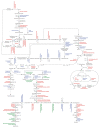
Results: A cDNA library was prepared for opium poppy cell cultures treated with a fungal elicitor for 10 h. Using 454 GS-FLX Titanium pyrosequencing, 427,369 expressed sequence tags (ESTs) with an average length of 462 bp were generated. Assembly of these sequences yielded 93,723 unigenes, of which 23,753 were assigned Gene Ontology annotations. Transcripts encoding all known sanguinarine biosynthetic enzymes were identified in the EST database, 5 of which were represented among the 50 most abundant transcripts. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) of total protein extracts from cell cultures treated with a fungal elicitor for 50 h facilitated the identification of 1,004 proteins. Proteins were fractionated by one-dimensional SDS-PAGE and digested with trypsin prior to LC-MS/MS analysis. Query of an opium poppy-specific EST database substantially enhanced peptide identification. Eight out of 10 known sanguinarine biosynthetic enzymes and many relevant primary metabolic enzymes were represented in the peptide database.
Conclusions: The integration of deep transcriptome and proteome analyses provides an effective platform to catalogue the components of secondary metabolism, and to identify genes encoding uncharacterized enzymes. The establishment of corresponding transcript and protein databases generated by next-generation technologies in a system with a well-defined metabolite profile facilitates an improved linkage between genes, enzymes, and pathway components. The proteome database represents the most relevant alkaloid-producing enzymes, compared with the much deeper and more complete transcriptome library. The transcript database contained full-length mRNAs encoding most alkaloid biosynthetic enzymes, which is a key requirement for the functional characterization of novel gene candidates.
Figures



References
-
- Zulak KG, Cornish A, Daskalchuk TE, Deyholos MK, Goodenowe DB, Gordon PP, Klassen D, Pelcher LE, Sensen CW, Facchini PJ. Gene transcript and metabolite profiling of elicitor-induced opium poppy cell cultures reveals the coordinate regulation of primary and secondary metabolism. Planta. 2007;225:1085–1106. doi: 10.1007/s00425-006-0419-5. - DOI - PubMed
-
- Zulak KG, Khan MF, Alcantara J, Schriemer DC, Facchini PJ. Plant defense responses in opium poppy cell cultures revealed by liquid chromatography tandem mass spectrometry proteomics. Molecular & Cellular Proteomics. 2009;8:86–98. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

