Characterization of a binding site for anionic phospholipids on KCNQ1
- PMID: 21084310
- PMCID: PMC3023506
- DOI: 10.1074/jbc.M110.153551
Characterization of a binding site for anionic phospholipids on KCNQ1
Abstract
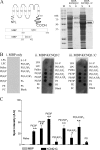
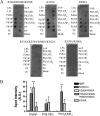
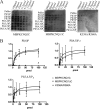
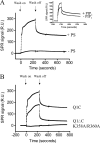
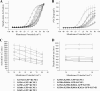
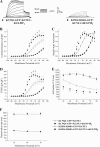
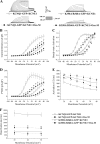
The KCNQ family of potassium channels underlie a repolarizing K(+) current in the heart and the M-current in neurones. The assembly of KCNQ1 with KCNE1 generates the delayed rectifier current I(Ks) in the heart. Characteristically these channels are regulated via G(q/11)-coupled receptors and the inhibition seen after phospholipase C activation is now thought to occur from membrane phosphatidylinositol (4,5)-bisphosphate (PIP(2)) depletion. It is not clear how KCNQ1 recognizes PIP(2) and specifically which residues in the channel complex are important. Using biochemical techniques we identify a cluster of basic residues namely, Lys-354, Lys-358, Arg-360, and Lys-362, in the proximal C terminus as being involved in binding anionic phospholipids. The mutation of specific residues in combination, to alanine leads to the loss of binding to phosphoinositides. Functionally, the introduction of these mutations into KCNQ1 leads to shifts in the voltage dependence of channel activation toward depolarized potentials and reductions in current density. Additionally, the biophysical effects of the charge neutralizing mutations, which disrupt phosphoinositide binding, mirror the effects we see on channel function when we deplete cellular PIP(2) levels through activation of a G(q/11)-coupled receptor. Conversely, the addition of diC8-PIP(2) to the wild-type channel, but not a PIP(2) binding-deficient mutant, acts to shift the voltage dependence of channel activation toward hyperpolarized potentials and increase current density. In conclusion, we use a combined biochemical and functional approach to identify a cluster of basic residues important for the binding and action of anionic phospholipids on the KCNQ1/KCNE1 complex.
Figures


Similar articles
-
Long QT mutations at the interface between KCNQ1 helix C and KCNE1 disrupt I(KS) regulation by PKA and PIP₂.J Cell Sci. 2014 Sep 15;127(Pt 18):3943-55. doi: 10.1242/jcs.147033. Epub 2014 Jul 18. J Cell Sci. 2014. PMID: 25037568 Free PMC article.
-
Mechanisms of disease pathogenesis in long QT syndrome type 5.Am J Physiol Cell Physiol. 2010 Feb;298(2):C263-73. doi: 10.1152/ajpcell.00308.2009. Epub 2009 Nov 11. Am J Physiol Cell Physiol. 2010. PMID: 19907016 Free PMC article.
-
KCNQ1 channels do not undergo concerted but sequential gating transitions in both the absence and the presence of KCNE1 protein.J Biol Chem. 2012 Oct 5;287(41):34212-24. doi: 10.1074/jbc.M112.364901. Epub 2012 Aug 20. J Biol Chem. 2012. PMID: 22908235 Free PMC article.
-
The I Ks Ion Channel Activator Mefenamic Acid Requires KCNE1 and Modulates Channel Gating in a Subunit-Dependent Manner.Mol Pharmacol. 2020 Feb;97(2):132-144. doi: 10.1124/mol.119.117952. Epub 2019 Nov 13. Mol Pharmacol. 2020. PMID: 31722973
-
Insights into Cardiac IKs (KCNQ1/KCNE1) Channels Regulation.Int J Mol Sci. 2020 Dec 11;21(24):9440. doi: 10.3390/ijms21249440. Int J Mol Sci. 2020. PMID: 33322401 Free PMC article. Review.
Cited by
-
Modulation of KV7 Channel Deactivation by PI(4,5)P2.Front Pharmacol. 2020 Jun 19;11:895. doi: 10.3389/fphar.2020.00895. eCollection 2020. Front Pharmacol. 2020. PMID: 32636742 Free PMC article.
-
Dual effect of phosphatidylinositol (4,5)-bisphosphate PIP(2) on Shaker K(+) [corrected] channels.J Biol Chem. 2012 Oct 19;287(43):36158-67. doi: 10.1074/jbc.M112.382085. Epub 2012 Aug 29. J Biol Chem. 2012. PMID: 22932893 Free PMC article.
-
Energetics and location of phosphoinositide binding in human Kir2.1 channels.J Biol Chem. 2013 Jun 7;288(23):16726-16737. doi: 10.1074/jbc.M113.452540. Epub 2013 Apr 5. J Biol Chem. 2013. PMID: 23564459 Free PMC article.
-
Long QT mutations at the interface between KCNQ1 helix C and KCNE1 disrupt I(KS) regulation by PKA and PIP₂.J Cell Sci. 2014 Sep 15;127(Pt 18):3943-55. doi: 10.1242/jcs.147033. Epub 2014 Jul 18. J Cell Sci. 2014. PMID: 25037568 Free PMC article.
-
Ca2+-Calmodulin and PIP2 interactions at the proximal C-terminus of Kv7 channels.Channels (Austin). 2017 Nov 2;11(6):686-695. doi: 10.1080/19336950.2017.1388478. Epub 2017 Nov 17. Channels (Austin). 2017. PMID: 28976808 Free PMC article.
References
-
- Barhanin J., Lesage F., Guillemare E., Fink M., Lazdunski M., Romey G. (1996) Nature 384, 78–80 - PubMed
-
- Sanguinetti M. C., Curran M. E., Zou A., Shen J., Spector P. S., Atkinson D. L., Keating M. T. (1996) Nature 384, 80–83 - PubMed
-
- Nerbonne J. M., Kass R. S. (2005) Physiol. Rev. 85, 1205–1253 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

