Mechanism of anthracycline-mediated down-regulation of GATA4 in the heart
- PMID: 21084315
- PMCID: PMC3058732
- DOI: 10.1093/cvr/cvq361
Mechanism of anthracycline-mediated down-regulation of GATA4 in the heart
Abstract
Aims: Anthracyclines such as daunorubicin (DNR) and doxorubicin are effective cancer chemotherapeutic agents, but can induce cardiotoxicity. GATA4 has been shown to serve as a survival factor of cardiac muscle cells, and anthracyclines promote apoptosis in part by down-regulating GATA4. The present study investigated the mechanism of anthracycline action to down-regulate GATA4.
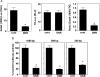
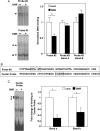
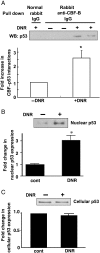
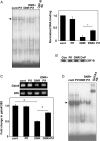
Methods and results: DNR inhibited the transcriptional activity exhibited by the 250 bp conserved region immediately upstream from the transcriptional start site of the Gata4 gene. Mapping this region identified that the CCAAT-binding factor/nuclear factor-Y (CBF/NF-Y) binding to the CCAAT box was inhibited by DNR in HL-1 cardiac muscle cells and in perfused isolated mouse hearts. The DNR action on the Gata4 promoter was found to be dependent on p53, since DNR promoted nuclear binding of p53 to CBF/NF-Y and pifithrin-α (a p53 inhibitor) attenuated DNR down-regulation of GATA4.
Conclusion: Anthracycline down-regulation of GATA4 is mediated by the inhibition of Gata4 gene transcription via a novel mechanism that involves the p53-dependent inhibition of CBF/NF-Y binding to the CCAAT box within the Gata4 promoter.
Figures


Similar articles
-
HSP27 regulates p53 transcriptional activity in doxorubicin-treated fibroblasts and cardiac H9c2 cells: p21 upregulation and G2/M phase cell cycle arrest.Am J Physiol Heart Circ Physiol. 2008 Apr;294(4):H1736-44. doi: 10.1152/ajpheart.91507.2007. Epub 2008 Feb 8. Am J Physiol Heart Circ Physiol. 2008. Retraction in: Am J Physiol Heart Circ Physiol. 2024 Aug 1;327(2):H445. doi: 10.1152/ajpheart.91507.2007_RET. PMID: 18263706 Retracted.
-
Pulmonary hypertension-induced GATA4 activation in the right ventricle.Hypertension. 2010 Dec;56(6):1145-51. doi: 10.1161/HYPERTENSIONAHA.110.160515. Epub 2010 Nov 8. Hypertension. 2010. PMID: 21059997 Free PMC article.
-
Disruption of a GATA4/Ankrd1 signaling axis in cardiomyocytes leads to sarcomere disarray: implications for anthracycline cardiomyopathy.PLoS One. 2012;7(4):e35743. doi: 10.1371/journal.pone.0035743. Epub 2012 Apr 20. PLoS One. 2012. PMID: 22532871 Free PMC article.
-
Atrial natriuretic peptide suppresses endothelin gene expression and proliferation in cardiac fibroblasts through a GATA4-dependent mechanism.Cardiovasc Res. 2009 Nov 1;84(2):209-17. doi: 10.1093/cvr/cvp208. Epub 2009 Jun 22. Cardiovasc Res. 2009. PMID: 19546173 Free PMC article.
-
Regulation of GATA4 transcriptional activity in cardiovascular development and disease.Curr Top Dev Biol. 2012;100:143-69. doi: 10.1016/B978-0-12-387786-4.00005-1. Curr Top Dev Biol. 2012. PMID: 22449843 Review.
Cited by
-
Phylogenetic origin of LI-cadherin revealed by protein and gene structure analysis.Cell Mol Life Sci. 2004 May;61(10):1157-66. doi: 10.1007/s00018-004-3470-z. Cell Mol Life Sci. 2004. PMID: 15141301 Free PMC article.
-
Melatonin and doxorubicin synergistically enhance apoptosis via autophagy-dependent reduction of AMPKα1 transcription in human breast cancer cells.Exp Mol Med. 2021 Sep;53(9):1413-1422. doi: 10.1038/s12276-021-00675-y. Epub 2021 Sep 28. Exp Mol Med. 2021. PMID: 34584194 Free PMC article.
-
MicroRNA-208a Silencing Attenuates Doxorubicin Induced Myocyte Apoptosis and Cardiac Dysfunction.Oxid Med Cell Longev. 2015;2015:597032. doi: 10.1155/2015/597032. Epub 2015 Jun 7. Oxid Med Cell Longev. 2015. PMID: 26137188 Free PMC article.
-
GATA4-targeted compound exhibits cardioprotective actions against doxorubicin-induced toxicity in vitro and in vivo: establishment of a chronic cardiotoxicity model using human iPSC-derived cardiomyocytes.Arch Toxicol. 2020 Jun;94(6):2113-2130. doi: 10.1007/s00204-020-02711-8. Epub 2020 Mar 17. Arch Toxicol. 2020. PMID: 32185414 Free PMC article.
-
The tell-tale heart: molecular and cellular responses to childhood anthracycline exposure.Am J Physiol Heart Circ Physiol. 2014 Nov 15;307(10):H1379-89. doi: 10.1152/ajpheart.00099.2014. Epub 2014 Sep 12. Am J Physiol Heart Circ Physiol. 2014. PMID: 25217655 Free PMC article. Review.
References
-
- Singal PK, Iliskovic N. Doxorubicin-induced cardiomyopathy. New Engl J Med. 1998;339:900–905. doi:10.1056/NEJM199809243391307. - DOI - PubMed
-
- Arola OJ, Saraste A, Pulkki K, Kallajoki M, Parvinen M, Voipio-Pulkki LM. Acute doxorubicin cardiotoxicity involves cardiomyocyte apoptosis. Cancer Res. 2000;60:1789–1792. - PubMed
-
- Kang YJ, Zhou ZX, Wang GW, Buridi A, Klein JB. Suppression by metallothionein of doxorubicin-induced cardiomyocyte apoptosis through inhibition of p38 mitogen-activated protein kinases. J Biol Chem. 2000;275:13690–13698. doi:10.1074/jbc.275.18.13690. - DOI - PubMed
-
- Suzuki YJ, Evans T. Regulation of cardiac myocyte apoptosis by the GATA-4 transcription factor. Life Sci. 2004;74:1829–1838. doi:10.1016/j.lfs.2003.10.002. - DOI - PubMed
-
- Kim Y, Ma AG, Kitta K, Fitch SN, Ikeda T, Ihara Y, et al. Anthracycline-induced suppression of GATA-4 transcription factor: implication in the regulation of cardiac myocyte apoptosis. Mol Pharmacol. 2003;63:368–377. doi:10.1124/mol.63.2.368. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

