The arginine clusters of the carboxy-terminal domain of the core protein of hepatitis B virus make pleiotropic contributions to genome replication
- PMID: 21084467
- PMCID: PMC3020508
- DOI: 10.1128/JVI.01957-10
The arginine clusters of the carboxy-terminal domain of the core protein of hepatitis B virus make pleiotropic contributions to genome replication
Abstract
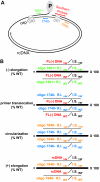
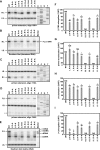
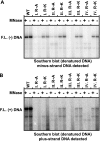
The carboxy-terminal domain (CTD) of the core protein of hepatitis B virus is not necessary for capsid assembly. However, the CTD does contribute to encapsidation of pregenomic RNA (pgRNA). The contribution of the CTD to DNA synthesis is less clear. This is the case because some mutations within the CTD increase the proportion of spliced RNA to pgRNA that are encapsidated and reverse transcribed. The CTD contains four clusters of consecutive arginine residues. The contributions of the individual arginine clusters to genome replication are unknown. We analyzed core protein variants in which the individual arginine clusters were substituted with either alanine or lysine residues. We developed assays to analyze these variants at specific steps throughout genome replication. We used a replication template that was not spliced in order to study the replication of only pgRNA. We found that alanine substitutions caused defects at both early and late steps in genome replication. Lysine substitutions also caused defects, but primarily during later steps. These findings demonstrate that the CTD contributes to DNA synthesis pleiotropically and that preserving the charge within the CTD is not sufficient to preserve function.
Figures

Similar articles
-
Phosphoacceptors threonine 162 and serines 170 and 178 within the carboxyl-terminal RRRS/T motif of the hepatitis B virus core protein make multiple contributions to hepatitis B virus replication.J Virol. 2014 Aug;88(16):8754-67. doi: 10.1128/JVI.01343-14. Epub 2014 May 21. J Virol. 2014. PMID: 24850741 Free PMC article.
-
Multiple roles of core protein linker in hepatitis B virus replication.PLoS Pathog. 2018 May 21;14(5):e1007085. doi: 10.1371/journal.ppat.1007085. eCollection 2018 May. PLoS Pathog. 2018. PMID: 29782550 Free PMC article.
-
Hepatitis B Virus Core Protein Dephosphorylation Occurs during Pregenomic RNA Encapsidation.J Virol. 2018 Jun 13;92(13):e02139-17. doi: 10.1128/JVI.02139-17. Print 2018 Jul 1. J Virol. 2018. PMID: 29669831 Free PMC article.
-
Phosphorylation of the Arginine-Rich C-Terminal Domains of the Hepatitis B Virus (HBV) Core Protein as a Fine Regulator of the Interaction between HBc and Nucleic Acid.Viruses. 2020 Jul 8;12(7):738. doi: 10.3390/v12070738. Viruses. 2020. PMID: 32650547 Free PMC article. Review.
-
Structure and function of the encapsidation signal of hepadnaviridae.J Viral Hepat. 1998 Nov;5(6):357-67. doi: 10.1046/j.1365-2893.1998.00124.x. J Viral Hepat. 1998. PMID: 9857345 Review.
Cited by
-
In vitro expression of precore proteins of hepatitis B virus subgenotype A1 is affected by HBcAg, and can affect HBsAg secretion.Sci Rep. 2021 Apr 14;11(1):8167. doi: 10.1038/s41598-021-87529-9. Sci Rep. 2021. PMID: 33854155 Free PMC article.
-
Immunoinformatics and Evaluation of Peptide Vaccines Derived from Global Hepatitis B Viral HBx and HBc Proteins Critical for Covalently Closed Circular DNA Integrity.Microorganisms. 2023 Nov 21;11(12):2826. doi: 10.3390/microorganisms11122826. Microorganisms. 2023. PMID: 38137971 Free PMC article.
-
Mutation of arginine residues to avoid non-specific cellular uptakes for hepatitis B virus core particles.J Nanobiotechnology. 2015 Feb 13;13:15. doi: 10.1186/s12951-015-0074-8. J Nanobiotechnology. 2015. PMID: 25890025 Free PMC article.
-
Hepatitis virus capsid polymorph stability depends on encapsulated cargo size.ACS Nano. 2013 Oct 22;7(10):8447-54. doi: 10.1021/nn4017839. Epub 2013 Sep 30. ACS Nano. 2013. PMID: 24010404 Free PMC article.
-
Peptidyl-prolyl cis/trans isomerase Pin1 interacts with hepatitis B virus core particle, but not with HBc protein, to promote HBV replication.Front Cell Infect Microbiol. 2023 Jun 19;13:1195063. doi: 10.3389/fcimb.2023.1195063. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37404723 Free PMC article.
References
-
- Auxilien, S., G. Keith, S. F. Le Grice, and J. L. Darlix. 1999. Role of posttranscriptional modifications of primer tRNALys,3 in the fidelity and efficacy of plus strand DNA transfer during HIV-1 reverse transcription. J. Biol. Chem. 274:4412-4420. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

