Bacterial HA1 vaccine against pandemic H5N1 influenza virus: evidence of oligomerization, hemagglutination, and cross-protective immunity in ferrets
- PMID: 21084473
- PMCID: PMC3020491
- DOI: 10.1128/JVI.02107-10
Bacterial HA1 vaccine against pandemic H5N1 influenza virus: evidence of oligomerization, hemagglutination, and cross-protective immunity in ferrets
Abstract
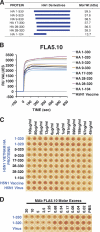
The impending influenza virus pandemic requires global vaccination to prevent large-scale mortality and morbidity, but traditional influenza virus vaccine production is too slow for rapid responses. We have developed bacterial systems for expression and purification of properly folded functional hemagglutinin as a rapid response to emerging pandemic strains. A recombinant H5N1 (A/Vietnam/1203/2004) hemagglutinin globular domain (HA1) was produced in Escherichia coli under controlled redox refolding conditions. Importantly, the properly folded HA1(1-320), i.e., HA1 lacking amino acids 321 to 330, contained ≥75% functional oligomers without addition of foreign oligomerization sequence. Site-directed mutagenesis mapped the oligomerization signal to the HA1 N-terminal Ile-Cys-Ile residues at positions 3 to 5. The purified HA1 oligomers (but not monomers) bound fetuin and agglutinated red blood cells. Upon immunization of rabbits, the oligomeric HA1(1-320) elicited potent neutralizing antibodies against homologous and heterologous H5N1 viruses more rapidly than HA1(28-320) containing only monomers. Ferrets vaccinated with oligomeric HA1 (but not monomeric HA1 with the N terminus deleted) at 15 and 3 μg/dose were fully protected from lethality and weight loss after challenge with homologous H5N1 (A/Vietnam/1203/2004, clade 1) virus, as well as heterologous clade 2.2 H5N1 (A/WooperSwan/Mongolia/244/2005) virus. Protection was associated with a significant reduction in viral loads in the nasal washes of homologous and heterologous virus challenged ferrets. This is the first study that describes the presence of an N-terminal oligomerization sequence in the globular domain of influenza virus hemagglutinin. Our findings suggest that functional oligomeric rHA1-based vaccines can be produced efficiently in bacterial systems and can be easily upscaled in response to a pandemic influenza virus threat.
Figures



Similar articles
-
H5 N-terminal β sheet promotes oligomerization of H7-HA1 that induces better antibody affinity maturation and enhanced protection against H7N7 and H7N9 viruses compared to inactivated influenza vaccine.Vaccine. 2014 Nov 12;32(48):6421-32. doi: 10.1016/j.vaccine.2014.09.049. Epub 2014 Oct 3. Vaccine. 2014. PMID: 25284811
-
Oligomeric recombinant H5 HA1 vaccine produced in bacteria protects ferrets from homologous and heterologous wild-type H5N1 influenza challenge and controls viral loads better than subunit H5N1 vaccine by eliciting high-affinity antibodies.J Virol. 2012 Nov;86(22):12283-93. doi: 10.1128/JVI.01596-12. Epub 2012 Sep 5. J Virol. 2012. PMID: 22951833 Free PMC article.
-
Flagellin-HA vaccines protect ferrets and mice against H5N1 highly pathogenic avian influenza virus (HPAIV) infections.Vaccine. 2012 Nov 6;30(48):6833-8. doi: 10.1016/j.vaccine.2012.09.013. Epub 2012 Sep 18. Vaccine. 2012. PMID: 23000130
-
Candidate influenza vaccines based on recombinant modified vaccinia virus Ankara.Expert Rev Vaccines. 2009 Apr;8(4):447-54. doi: 10.1586/erv.09.4. Expert Rev Vaccines. 2009. PMID: 19348560 Free PMC article. Review.
-
Advances and future challenges in recombinant adenoviral vectored H5N1 influenza vaccines.Viruses. 2012 Nov 1;4(11):2711-35. doi: 10.3390/v4112711. Viruses. 2012. PMID: 23202501 Free PMC article. Review.
Cited by
-
Development of a universal CTL-based vaccine for influenza.Bioengineered. 2013 Nov-Dec;4(6):374-8. doi: 10.4161/bioe.23573. Epub 2013 Jan 21. Bioengineered. 2013. PMID: 23337287 Free PMC article. Review.
-
Antigenic and immunogenic properties of recombinant hemagglutinin proteins from H1N1 A/Brisbane/59/07 and B/Florida/04/06 when produced in various protein expression systems.Vaccine. 2012 Jun 29;30(31):4606-16. doi: 10.1016/j.vaccine.2012.05.005. Epub 2012 May 15. Vaccine. 2012. PMID: 22609035 Free PMC article.
-
Decreased serologic response in vaccinated military recruits during 2011 correspond to genetic drift in concurrent circulating pandemic A/H1N1 viruses.PLoS One. 2012;7(4):e34581. doi: 10.1371/journal.pone.0034581. Epub 2012 Apr 13. PLoS One. 2012. PMID: 22514639 Free PMC article.
-
MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines.Sci Transl Med. 2011 Jun 1;3(85):85ra48. doi: 10.1126/scitranslmed.3002336. Sci Transl Med. 2011. PMID: 21632986 Free PMC article. Clinical Trial.
-
Glycosylation of the HIV-1 Env V1V2 loop to form a native-like structure may not be essential with a nanoparticle vaccine.Future Virol. 2019 Feb;14(2):51-54. doi: 10.2217/fvl-2018-0174. Epub 2019 Jan 10. Future Virol. 2019. PMID: 30815025 Free PMC article. No abstract available.
References
-
- Bizebard, T., et al. 1995. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature 376:92-94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

