To polyadenylate or to deadenylate: that is the question
- PMID: 21084869
- PMCID: PMC3048043
- DOI: 10.4161/cc.9.22.13887
To polyadenylate or to deadenylate: that is the question
Abstract
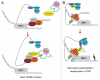
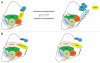
mRNA polyadenylation and deadenylation are important processes that allow rapid regulation of gene expression in response to different cellular conditions. Almost all eukaryotic mRNA precursors undergo a co-transcriptional cleavage followed by polyadenylation at the 3' end. After the signals are selected, polyadenylation occurs to full extent, suggesting that this first round of polyadenylation is a default modification for most mRNAs. However, the length of these poly(A) tails changes by the activation of deadenylation, which might regulate gene expression by affecting mRNA stability, mRNA transport, or translation initiation. The mechanisms behind deadenylation activation are highly regulated and associated with cellular conditions such as development, mRNA surveillance, DNA damage response, cell differentiation and cancer. After deadenylation, depending on the cellular response, some mRNAs might undergo an extension of the poly(A) tail or degradation. The polyadenylation/deadenylation machinery itself, miRNAs, or RNA binding factors are involved in the regulation of polyadenylation/deadenylation. Here, we review the mechanistic connections between polyadenylation and deadenylation and how the two processes are regulated in different cellular conditions. It is our conviction that further studies of the interplay between polyadenylation and deadenylation will provide critical information required for a mechanistic understanding of several diseases, including cancer development.
Figures


References
-
- Bentley DL. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. - PubMed
-
- Sheets MD, Wickens M. Two phases in the addition of a poly(A) tail. Genes Dev. 1989;3:1401–1412. - PubMed
-
- Gilmartin GM. Eukaryotic mRNA 3′ processing: A common means to different ends. Genes Dev. 2005;19:2517–2521. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
