Cap binding and immune evasion revealed by Lassa nucleoprotein structure
- PMID: 21085117
- PMCID: PMC3057469
- DOI: 10.1038/nature09605
Cap binding and immune evasion revealed by Lassa nucleoprotein structure
Abstract
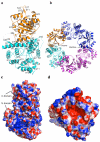
Lassa virus, the causative agent of Lassa fever, causes thousands of deaths annually and is a biological threat agent, for which there is no vaccine and limited therapy. The nucleoprotein (NP) of Lassa virus has essential roles in viral RNA synthesis and immune suppression, the molecular mechanisms of which are poorly understood. Here we report the crystal structure of Lassa virus NP at 1.80 Å resolution, which reveals amino (N)- and carboxy (C)-terminal domains with structures unlike any of the reported viral NPs. The N domain folds into a novel structure with a deep cavity for binding the m7GpppN cap structure that is required for viral RNA transcription, whereas the C domain contains 3'-5' exoribonuclease activity involved in suppressing interferon induction. To our knowledge this is the first X-ray crystal structure solved for an arenaviral NP, which reveals its unexpected functions and indicates unique mechanisms in cap binding and immune evasion. These findings provide great potential for vaccine and drug development.
Figures


Comment in
-
Virology: One protein, many functions.Nature. 2010 Dec 9;468(7325):773-5. doi: 10.1038/468773a. Nature. 2010. PMID: 21150990 No abstract available.
Similar articles
-
Virology: One protein, many functions.Nature. 2010 Dec 9;468(7325):773-5. doi: 10.1038/468773a. Nature. 2010. PMID: 21150990 No abstract available.
-
Structures of arenaviral nucleoproteins with triphosphate dsRNA reveal a unique mechanism of immune suppression.J Biol Chem. 2013 Jun 7;288(23):16949-16959. doi: 10.1074/jbc.M112.420521. Epub 2013 Apr 24. J Biol Chem. 2013. PMID: 23615902 Free PMC article.
-
Insight into the binding modes of Lassa nucleoprotein complexed with ssRNA by molecular dynamic simulations and free energy calculations.J Biomol Struct Dyn. 2015;33(5):946-60. doi: 10.1080/07391102.2014.923785. Epub 2014 Jun 19. J Biomol Struct Dyn. 2015. PMID: 24824824
-
Influenza virus nucleoprotein: structure, RNA binding, oligomerization and antiviral drug target.Future Microbiol. 2013 Dec;8(12):1537-45. doi: 10.2217/fmb.13.128. Future Microbiol. 2013. PMID: 24266354 Review.
-
Hiding the evidence: two strategies for innate immune evasion by hemorrhagic fever viruses.Curr Opin Virol. 2012 Apr;2(2):151-6. doi: 10.1016/j.coviro.2012.01.003. Epub 2012 Jan 28. Curr Opin Virol. 2012. PMID: 22482712 Free PMC article. Review.
Cited by
-
Lymphocytic Choriomeningitis Virus Differentially Affects the Virus-Induced Type I Interferon Response and Mitochondrial Apoptosis Mediated by RIG-I/MAVS.J Virol. 2015 Jun;89(12):6240-50. doi: 10.1128/JVI.00610-15. Epub 2015 Apr 1. J Virol. 2015. PMID: 25833049 Free PMC article.
-
Multifunctional nature of the arenavirus RING finger protein Z.Viruses. 2012 Nov 9;4(11):2973-3011. doi: 10.3390/v4112973. Viruses. 2012. PMID: 23202512 Free PMC article. Review.
-
Brothers in Arms: Structure, Assembly and Function of Arenaviridae Nucleoprotein.Viruses. 2020 Jul 17;12(7):772. doi: 10.3390/v12070772. Viruses. 2020. PMID: 32708976 Free PMC article. Review.
-
Identification of virulence determinants within the L genomic segment of the pichinde arenavirus.J Virol. 2013 Jun;87(12):6635-43. doi: 10.1128/JVI.00044-13. Epub 2013 Apr 3. J Virol. 2013. PMID: 23552411 Free PMC article.
-
Suppression of the interferon and NF-κB responses by severe fever with thrombocytopenia syndrome virus.J Virol. 2012 Aug;86(16):8388-401. doi: 10.1128/JVI.00612-12. Epub 2012 May 23. J Virol. 2012. PMID: 22623799 Free PMC article.
References
-
- Buchmeier MJ, De La Torre JC, Peters CJ. In: Fields Virology. Knipe DM, Howley PM, editors. Vol. 2. Lippincott Williams & Wilkins; 2007. pp. 1791–1827.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
- R01 AI067704/AI/NIAID NIH HHS/United States
- R01 AI083409/AI/NIAID NIH HHS/United States
- U19 AI057266/AI/NIAID NIH HHS/United States
- 5-U19-AI057266-07/AI/NIAID NIH HHS/United States
- AI067704/AI/NIAID NIH HHS/United States
- DK64399/DK/NIDDK NIH HHS/United States
- 083501/Z/07/Z/WT_/Wellcome Trust/United Kingdom
- R24 DK064399/DK/NIDDK NIH HHS/United States
- 5-U54-AI-057157-06/AI/NIAID NIH HHS/United States
- R01 AI093580/AI/NIAID NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01AI083409/AI/NIAID NIH HHS/United States
- U54 AI057157/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

