Quantitative reactivity profiling predicts functional cysteines in proteomes
- PMID: 21085121
- PMCID: PMC3058684
- DOI: 10.1038/nature09472
Quantitative reactivity profiling predicts functional cysteines in proteomes
Abstract
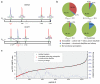
Cysteine is the most intrinsically nucleophilic amino acid in proteins, where its reactivity is tuned to perform diverse biochemical functions. The absence of a consensus sequence that defines functional cysteines in proteins has hindered their discovery and characterization. Here we describe a proteomics method to profile quantitatively the intrinsic reactivity of cysteine residues en masse directly in native biological systems. Hyper-reactivity was a rare feature among cysteines and it was found to specify a wide range of activities, including nucleophilic and reductive catalysis and sites of oxidative modification. Hyper-reactive cysteines were identified in several proteins of uncharacterized function, including a residue conserved across eukaryotic phylogeny that we show is required for yeast viability and is involved in iron-sulphur protein biogenesis. We also demonstrate that quantitative reactivity profiling can form the basis for screening and functional assignment of cysteines in computationally designed proteins, where it discriminated catalytically active from inactive cysteine hydrolase designs.
Figures
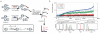
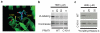


References
-
- Eisenberg D, Marcotte EM, Xenarios I, Yeates TO. Protein function in the post-genomic era. Nature. 2000;405:823–826. - PubMed
-
- Bulaj G, Kortemme T, Goldenberg DP. Ionization reactivity relationships for cysteine thiols in polypeptides. Biochemistry. 1998;37:8965–8972. - PubMed
-
- Giles NM, Giles GI, Jacob C. Multiple roles of cysteine in biocatalysis. Biochem. Biophys. Res. Commun. 2003;300:1–4. - PubMed
-
- Reddie KG, Carroll KS. Expanding the functional diversity of proteins through cysteine oxidation. Curr. Opin. Chem. Biol. 2008;12:746–754. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

