Interaction of the retinoblastoma protein with Orc1 and its recruitment to human origins of DNA replication
- PMID: 21085491
- PMCID: PMC2976706
- DOI: 10.1371/journal.pone.0013720
Interaction of the retinoblastoma protein with Orc1 and its recruitment to human origins of DNA replication
Abstract
Background: The retinoblastoma protein (Rb) is a crucial regulator of cell cycle progression by binding with E2F transcription factor and repressing the expression of a variety of genes required for the G1-S phase transition.
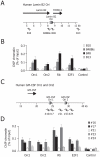
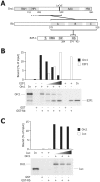
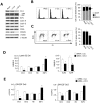
Methodology/principal findings: Here we show that Rb and E2F1 directly participate in the control of initiation of DNA replication in human HeLa, U2OS and T98G cells by specifically binding to origins of DNA replication in a cell cycle regulated manner. We show that, both in vitro and inside the cells, the largest subunit of the origin recognition complex (Orc1) specifically binds hypo-phosphorylated Rb and that this interaction is competitive with the binding of Rb to E2F1. The displacement of Rb-bound Orc1 by E2F1 at origins of DNA replication marks the progression of the G1 phase of the cell cycle toward the G1-S border.
Conclusions/significance: The participation of Rb and E2F1 in the formation of the multiprotein complex that binds origins of DNA replication in mammalian cells appears to represent an effective mechanism to couple the expression of genes required for cell cycle progression to the activation of DNA replication.
Conflict of interest statement
Figures




Similar articles
-
Orc1 Binding to Mitotic Chromosomes Precedes Spatial Patterning during G1 Phase and Assembly of the Origin Recognition Complex in Human Cells.J Biol Chem. 2015 May 8;290(19):12355-69. doi: 10.1074/jbc.M114.625012. Epub 2015 Mar 17. J Biol Chem. 2015. PMID: 25784553 Free PMC article.
-
Origin licensing and p53 status regulate Cdk2 activity during G(1).Cell Cycle. 2009 Jun 15;8(12):1952-63. doi: 10.4161/cc.8.12.8811. Epub 2009 Jun 21. Cell Cycle. 2009. PMID: 19440053 Free PMC article.
-
Genome-wide mapping of human DNA-replication origins: levels of transcription at ORC1 sites regulate origin selection and replication timing.Genome Res. 2013 Jan;23(1):1-11. doi: 10.1101/gr.142331.112. Epub 2012 Nov 27. Genome Res. 2013. PMID: 23187890 Free PMC article.
-
Cyclin-dependent kinases and S phase control in mammalian cells.Cell Cycle. 2003 Jul-Aug;2(4):316-24. Cell Cycle. 2003. PMID: 12851482 Review.
-
[Regulation of initiation of DNA replication in through G1 to S phases: overview].Tanpakushitsu Kakusan Koso. 2009 Mar;54(4 Suppl):317-9. Tanpakushitsu Kakusan Koso. 2009. PMID: 21089469 Review. Japanese. No abstract available.
Cited by
-
A retinoblastoma allele that is mutated at its common E2F interaction site inhibits cell proliferation in gene-targeted mice.Mol Cell Biol. 2014 Jun;34(11):2029-45. doi: 10.1128/MCB.01589-13. Epub 2014 Mar 24. Mol Cell Biol. 2014. PMID: 24662053 Free PMC article.
-
Hippo signaling is intrinsically regulated during cell cycle progression by APC/CCdh1.Proc Natl Acad Sci U S A. 2019 May 7;116(19):9423-9432. doi: 10.1073/pnas.1821370116. Epub 2019 Apr 18. Proc Natl Acad Sci U S A. 2019. PMID: 31000600 Free PMC article.
-
Regulating DNA replication in eukarya.Cold Spring Harb Perspect Biol. 2013 Sep 1;5(9):a012930. doi: 10.1101/cshperspect.a012930. Cold Spring Harb Perspect Biol. 2013. PMID: 23838438 Free PMC article. Review.
-
The Replicative Consequences of Papillomavirus E2 Protein Binding to the Origin Replication Factor ORC2.PLoS Pathog. 2016 Oct 4;12(10):e1005934. doi: 10.1371/journal.ppat.1005934. eCollection 2016 Oct. PLoS Pathog. 2016. PMID: 27701460 Free PMC article.
-
The SMC5/6 Complex Represses the Replicative Program of High-Risk Human Papillomavirus Type 31.Pathogens. 2020 Sep 25;9(10):786. doi: 10.3390/pathogens9100786. Pathogens. 2020. PMID: 32992873 Free PMC article.
References
-
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. - PubMed
-
- Diffley JF. Regulation of early events in chromosome replication. Curr Biol. 2004;14:R778–786. - PubMed
-
- Giacca M, Falaschi A. Specification of DNA replication origins of mammalian genomes. Chemtracts. 2002;15:519–532.
-
- Stevaux O, Dyson NJ. A revised picture of the E2F transcriptional network and RB function. Curr Opin Cell Biol. 2002;14:684–691. - PubMed
-
- Tamrakar S, Rubin E, Ludlow JW. Role of pRB dephosphorylation in cell cycle regulation. Front Biosci. 2000;5:D121–D137. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

