Extensive co-operation between the Epstein-Barr virus EBNA3 proteins in the manipulation of host gene expression and epigenetic chromatin modification
- PMID: 21085583
- PMCID: PMC2981562
- DOI: 10.1371/journal.pone.0013979
Extensive co-operation between the Epstein-Barr virus EBNA3 proteins in the manipulation of host gene expression and epigenetic chromatin modification
Abstract
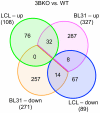
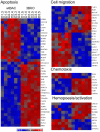
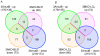
Epstein-Barr virus (EBV) is able to drive the transformation of B-cells, resulting in the generation of lymphoblastoid cell lines (LCLs) in vitro. EBV nuclear proteins EBNA3A and EBNA3C are necessary for efficient transformation, while EBNA3B is dispensable. We describe a transcriptome analysis of BL31 cells infected with a series of EBNA3-knockout EBVs, including one deleted for all three EBNA3 genes. Using Affymetrix Exon 1.0 ST microarrays analysed with the MMBGX algorithm, we have identified over 1000 genes whose regulation by EBV requires one of the EBNA3s. Remarkably, a third of the genes identified require more than one EBNA3 for their regulation, predominantly EBNA3C co-operating with either EBNA3B, EBNA3A or both. The microarray was validated by real-time PCR, while ChIP analysis of a selection of co-operatively repressed promoters indicates a role for polycomb group complexes. Targets include genes involved in apoptosis, cell migration and B-cell differentiation, and show a highly significant but subtle alteration in genes involved in mitosis. In order to assess the relevance of the BL31 system to LCLs, we analysed the transcriptome of a set of EBNA3B knockout (3BKO) LCLs. Around a third of the genes whose expression level in LCLs was altered in the absence of EBNA3B were also altered in 3BKO-BL31 cell lines.Among these are TERT and TCL1A, implying that EBV-induced changes in the expression of these genes are not required for B-cell transformation. We also identify 26 genes that require both EBNA3A and EBNA3B for their regulation in LCLs. Together, this shows the complexity of the interaction between EBV and its host, whereby multiple EBNA3 proteins co-operate to modulate the behaviour of the host cell.
Conflict of interest statement
Figures

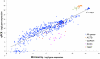

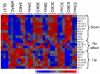

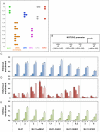
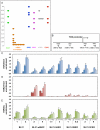
References
-
- Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. - PubMed
-
- Thorley-Lawson DA, Gross A. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. The New England Journal of Medicine. 2004;350:1328–1337. - PubMed
-
- Thorley-Lawson DA, Allday MJ. The curious case of the tumour virus: 50 years of Burkitt's lymphoma. Nat Rev Microbiol. 2008;6:913–924. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

