Whole genome sequencing highlights genetic changes associated with laboratory domestication of C. elegans
- PMID: 21085631
- PMCID: PMC2978686
- DOI: 10.1371/journal.pone.0013922
Whole genome sequencing highlights genetic changes associated with laboratory domestication of C. elegans
Abstract
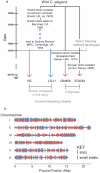
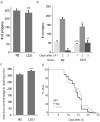
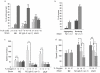
Defining the mutational landscape when individuals of a species grow separately and diverge over many generations can provide insights into trait evolution. A specific example of this involves studying changes associated with domestication where different lines of the same wild stock have been cultivated independently in different standard environments. Whole genome sequence comparison of such lines permits estimation of mutation rates, inference of genes' ancestral states and ancestry of existing strains, and correction of sequencing errors in genome databases. Here we study domestication of the C. elegans Bristol strain as a model, and report the genome sequence of LSJ1 (Bristol), a sibling of the standard C. elegans reference wild type N2 (Bristol). The LSJ1 and N2 lines were cultivated separately from shortly after the Bristol strain was isolated until methods to freeze C. elegans were developed. We find that during this time the two strains have accumulated 1208 genetic differences. We describe phenotypic variation between N2 and LSJ1 in the rate at which embryos develop, the rate of production of eggs, the maturity of eggs at laying, and feeding behavior, all the result of post-isolation changes. We infer the ancestral alleles in the original Bristol isolate and highlight 2038 likely sequencing errors in the original N2 reference genome sequence. Many of these changes modify genome annotation. Our study provides a starting point to further investigate genotype-phenotype association and offers insights into the process of selection as a result of laboratory domestication.
Conflict of interest statement
Figures

Similar articles
-
C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy.PLoS One. 2010 Nov 8;5(11):e15435. doi: 10.1371/journal.pone.0015435. PLoS One. 2010. PMID: 21079745 Free PMC article.
-
Punctuated Loci on Chromosome IV Determine Natural Variation in Orsay Virus Susceptibility of Caenorhabditis elegans Strains Bristol N2 and Hawaiian CB4856.J Virol. 2021 May 24;95(12):e02430-20. doi: 10.1128/JVI.02430-20. Print 2021 May 24. J Virol. 2021. PMID: 33827942 Free PMC article.
-
A genome-wide view of Caenorhabditis elegans base-substitution mutation processes.Proc Natl Acad Sci U S A. 2009 Sep 22;106(38):16310-4. doi: 10.1073/pnas.0904895106. Epub 2009 Sep 10. Proc Natl Acad Sci U S A. 2009. PMID: 19805298 Free PMC article.
-
Comparative genomics in C. elegans, C. briggsae, and other Caenorhabditis species.Methods Mol Biol. 2006;351:13-29. doi: 10.1385/1-59745-151-7:13. Methods Mol Biol. 2006. PMID: 16988423 Review.
-
Genome evolution in Caenorhabditis.Brief Funct Genomic Proteomic. 2008 May;7(3):211-6. doi: 10.1093/bfgp/eln022. Epub 2008 Jun 23. Brief Funct Genomic Proteomic. 2008. PMID: 18573804 Review.
Cited by
-
The laboratory domestication of Caenorhabditis elegans.Trends Genet. 2015 May;31(5):224-31. doi: 10.1016/j.tig.2015.02.009. Epub 2015 Mar 21. Trends Genet. 2015. PMID: 25804345 Free PMC article. Review.
-
Experimental Evolution with Caenorhabditis Nematodes.Genetics. 2017 Jun;206(2):691-716. doi: 10.1534/genetics.115.186288. Genetics. 2017. PMID: 28592504 Free PMC article. Review.
-
Reproductive consequences of transient pathogen exposure across host genotypes and generations.Ecol Evol. 2022 Mar 21;12(3):e8720. doi: 10.1002/ece3.8720. eCollection 2022 Mar. Ecol Evol. 2022. PMID: 35356553 Free PMC article.
-
Nematode gene annotation by machine-learning-assisted proteotranscriptomics enables proteome-wide evolutionary analysis.Genome Res. 2023 Jan;33(1):112-128. doi: 10.1101/gr.277070.122. Epub 2023 Jan 18. Genome Res. 2023. PMID: 36653121 Free PMC article.
-
Genome-wide variations in a natural isolate of the nematode Caenorhabditis elegans.BMC Genomics. 2014 Apr 2;15:255. doi: 10.1186/1471-2164-15-255. BMC Genomics. 2014. PMID: 24694239 Free PMC article.
References
-
- Darwin C. London: John Murray; 1875. The variation of animals and plants under domestication.
-
- Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–707. - PubMed
-
- Bridges C, Brehme K. Washington, D.C.: Carnegie Institute of Washington; 1944. The mutants of Drosophila melanogaster.257
-
- Lindsley D, Grell E. Washington, D.C.: Carnegie Institute of Washington.; 1968. Genetic variations of Drosophila melanogaster.469
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

