Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch
- PMID: 21085632
- PMCID: PMC2978687
- DOI: 10.1371/journal.pgen.1001205
Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch
Abstract
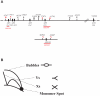
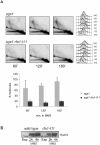
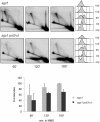
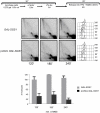
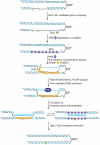
Damage tolerance mechanisms mediating damage-bypass and gap-filling are crucial for genome integrity. A major damage tolerance pathway involves recombination and is referred to as template switch. Template switch intermediates were visualized by 2D gel electrophoresis in the proximity of replication forks as X-shaped structures involving sister chromatid junctions. The homologous recombination factor Rad51 is required for the formation/stabilization of these intermediates, but its mode of action remains to be investigated. By using a combination of genetic and physical approaches, we show that the homologous recombination factors Rad55 and Rad57, but not Rad59, are required for the formation of template switch intermediates. The replication-proficient but recombination-defective rfa1-t11 mutant is normal in triggering a checkpoint response following DNA damage but is impaired in X-structure formation. The Exo1 nuclease also has stimulatory roles in this process. The checkpoint kinase, Rad53, is required for X-molecule formation and phosphorylates Rad55 robustly in response to DNA damage. Although Rad55 phosphorylation is thought to activate recombinational repair under conditions of genotoxic stress, we find that Rad55 phosphomutants do not affect the efficiency of X-molecule formation. We also examined the DNA polymerase implicated in the DNA synthesis step of template switch. Deficiencies in translesion synthesis polymerases do not affect X-molecule formation, whereas DNA polymerase δ, required also for bulk DNA synthesis, plays an important role. Our data indicate that a subset of homologous recombination factors, together with DNA polymerase δ, promote the formation of template switch intermediates that are then preferentially dissolved by the action of the Sgs1 helicase in association with the Top3 topoisomerase rather than resolved by Holliday Junction nucleases. Our results allow us to propose the choreography through which different players contribute to template switch in response to DNA damage and to distinguish this process from other recombination-mediated processes promoting DNA repair.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. - PubMed
-
- Lehmann AR, Fuchs RP. Gaps and forks in DNA replication: Rediscovering old models. DNA Repair (Amst) 2006;5:1495–1498. - PubMed
-
- Friedberg EC. Suffering in silence: the tolerance of DNA damage. Nat Rev Mol Cell Biol. 2005;6:943–953. - PubMed
-
- Higgins NP, Kato K, Strauss B. A model for replication repair in mammalian cells. J Mol Biol. 1976;101:417–425. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

