Local gene regulation details a recognition code within the LacI transcriptional factor family
- PMID: 21085639
- PMCID: PMC2978694
- DOI: 10.1371/journal.pcbi.1000989
Local gene regulation details a recognition code within the LacI transcriptional factor family
Abstract
The specific binding of regulatory proteins to DNA sequences exhibits no clear patterns of association between amino acids (AAs) and nucleotides (NTs). This complexity of protein-DNA interactions raises the question of whether a simple set of wide-coverage recognition rules can ever be identified. Here, we analyzed this issue using the extensive LacI family of transcriptional factors (TFs). We searched for recognition patterns by introducing a new approach to phylogenetic footprinting, based on the pervasive presence of local regulation in prokaryotic transcriptional networks. We identified a set of specificity correlations--determined by two AAs of the TFs and two NTs in the binding sites--that is conserved throughout a dominant subgroup within the family regardless of the evolutionary distance, and that act as a relatively consistent recognition code. The proposed rules are confirmed with data of previous experimental studies and by events of convergent evolution in the phylogenetic tree. The presence of a code emphasizes the stable structural context of the LacI family, while defining a precise blueprint to reprogram TF specificity with many practical applications.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
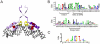
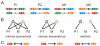
 in the sense strand of the palindromic combinations.
in the sense strand of the palindromic combinations.
 . We included the case for (AA-15, AA-16) = YQ corresponding to the synthetic SymL site in C). Recognition degeneracies are represented as unidirectional arrows (asymmetrical intrinsic), bidirectional divergent arrows (symmetrical intrinsic), and bidirectional convergent arrows (extrinsic). Colors for polar (green), basic (blue), acidic (red) and hydrophobic (black) amino acids. B) Agreement between synthetic and natural data. Recognition of (NT-5, NT-4)-palindromes by different (AA-15, AA-16)-LacI mutants (YQ is the wild type). Data from –from which we only considered those sequences (AA-15, AA-16) with a natural correspondence in Table S1. Rest of BS positions as in SymL. The larger the TF/BS affinity, the stronger the repression of the
. We included the case for (AA-15, AA-16) = YQ corresponding to the synthetic SymL site in C). Recognition degeneracies are represented as unidirectional arrows (asymmetrical intrinsic), bidirectional divergent arrows (symmetrical intrinsic), and bidirectional convergent arrows (extrinsic). Colors for polar (green), basic (blue), acidic (red) and hydrophobic (black) amino acids. B) Agreement between synthetic and natural data. Recognition of (NT-5, NT-4)-palindromes by different (AA-15, AA-16)-LacI mutants (YQ is the wild type). Data from –from which we only considered those sequences (AA-15, AA-16) with a natural correspondence in Table S1. Rest of BS positions as in SymL. The larger the TF/BS affinity, the stronger the repression of the  -galactosidase activity. Experimental conditions limited repression to a factor of 200. Arrows indicated again degeneracy classes. Predictions for wild type YQ correspond to asymmetric natural BSs (see text). (NT-5, NT-4)-palindromes involved in the predicted correlations for PM (
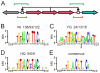
-galactosidase activity. Experimental conditions limited repression to a factor of 200. Arrows indicated again degeneracy classes. Predictions for wild type YQ correspond to asymmetric natural BSs (see text). (NT-5, NT-4)-palindromes involved in the predicted correlations for PM ( , see Table S1) lack an experimental test. Accordingly, PM do not exhibit a strong affinity for any of the tested palindromes (see Fig. S3), C) Natural and synthetic operators. A dot distinguishes the half sites. Flanking nucleotides separated by a space to help visualization of the highly conserved central region of the BSs. Colors identify different palindromic or mixed combinations in the specificity nucleotides (see Table S2 for more details).
, see Table S1) lack an experimental test. Accordingly, PM do not exhibit a strong affinity for any of the tested palindromes (see Fig. S3), C) Natural and synthetic operators. A dot distinguishes the half sites. Flanking nucleotides separated by a space to help visualization of the highly conserved central region of the BSs. Colors identify different palindromic or mixed combinations in the specificity nucleotides (see Table S2 for more details).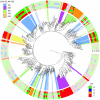
 binding to natural SymL-like BSs (Fig. 4.C and Table S2). Only one BS per TF is shown. The external color code displays the specificity-associated positions –to help visualization of palindromic combinations right positions are read in the complementary (c) strand:
binding to natural SymL-like BSs (Fig. 4.C and Table S2). Only one BS per TF is shown. The external color code displays the specificity-associated positions –to help visualization of palindromic combinations right positions are read in the complementary (c) strand:  . The color background in several branches corresponds to different recognition AAs (only a few recognition classes were enhanced). External color code in these branches shows darker colors to help visualization. Dots in branches denote bootstrap values larger than 80 (for 100 trees total, see Fig. S4 for more details).
. The color background in several branches corresponds to different recognition AAs (only a few recognition classes were enhanced). External color code in these branches shows darker colors to help visualization. Dots in branches denote bootstrap values larger than 80 (for 100 trees total, see Fig. S4 for more details).References
-
- Pabo CO, Sauer RT. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. - PubMed
-
- Suzuki M, Brenner SE, Gerstein M, Yagi N. DNA recognition code of transcription factors. Protein Eng Des Sel. 1995;8:319–328. - PubMed
-
- Choo Y, Klug A. Physical basis of a protein-DNA recognition code. Current Opinion In Struct Biol. 1997;7:117–125. - PubMed

