Quantitative assessment of the sensitivity of various commercial reverse transcriptases based on armored HIV RNA
- PMID: 21085668
- PMCID: PMC2978101
- DOI: 10.1371/journal.pone.0013931
Quantitative assessment of the sensitivity of various commercial reverse transcriptases based on armored HIV RNA
Abstract
Background: The in-vitro reverse transcription of RNA to its complementary DNA, catalyzed by the enzyme reverse transcriptase, is the most fundamental step in the quantitative RNA detection in genomic studies. As such, this step should be as analytically sensitive, efficient and reproducible as possible, especially when dealing with degraded or low copy RNA samples. While there are many reverse transcriptases in the market, all claiming to be highly sensitive, there is need for a systematic independent comparison of their applicability in quantification of rare RNA transcripts or low copy RNA, such as those obtained from archival tissues.
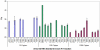
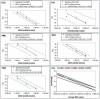
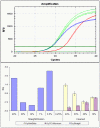
Methodology/principal findings: We performed RT-qPCR to assess the sensitivity and reproducibility of 11 commercially available reverse transcriptases in cDNA synthesis from low copy number RNA levels. As target RNA, we used a serially known number of Armored HIV RNA molecules, and observed that 9 enzymes we tested were consistently sensitive to ∼1,000 copies, seven of which were sensitive to ∼100 copies, while only 5 were sensitive to ∼10 RNA template copies across all replicates tested. Despite their demonstrated sensitivity, these five best performing enzymes (Accuscript, HIV-RT, M-MLV, Superscript III and Thermoscript) showed considerable variation in their reproducibility as well as their overall amplification efficiency. Accuscript and Superscript III were the most sensitive and consistent within runs, with Accuscript and Superscript II ranking as the most reproducible enzymes between assays.
Conclusions/significance: We therefore recommend the use of Accuscript or Superscript III when dealing with low copy number RNA levels, and suggest purification of the RT reactions prior to downstream applications (eg qPCR) to augment detection. Although the results presented in this study were based on a viral RNA surrogate, and applied to nucleic acid lysates derived from archival formalin-fixed paraffin embedded tissue, their relative performance on RNA obtained from other tissue types may vary, and needs future evaluation.
Conflict of interest statement
Figures
Similar articles
-
Variables influencing the efficiency and interpretation of reverse transcription quantitative PCR (RT-qPCR): An empirical study using Bacteriophage MS2.J Virol Methods. 2017 Mar;241:1-10. doi: 10.1016/j.jviromet.2016.12.002. Epub 2016 Dec 7. J Virol Methods. 2017. PMID: 27940257
-
A M-MLV reverse transcriptase with reduced RNaseH activity allows greater sensitivity of gene expression detection in formalin fixed and paraffin embedded prostate cancer samples.Exp Mol Pathol. 2013 Aug;95(1):98-104. doi: 10.1016/j.yexmp.2013.05.008. Epub 2013 Jun 2. Exp Mol Pathol. 2013. PMID: 23739432
-
Performance Comparison of Reverse Transcriptases for Single-Cell Studies.Clin Chem. 2020 Jan 1;66(1):217-228. doi: 10.1373/clinchem.2019.307835. Clin Chem. 2020. PMID: 31699702
-
Amplification of cDNA Generated by Reverse Transcription of mRNA: Two-Step Reverse Transcription-Polymerase Chain Reaction (RT-PCR).Cold Spring Harb Protoc. 2019 May 1;2019(5). doi: 10.1101/pdb.prot095190. Cold Spring Harb Protoc. 2019. PMID: 31043555
-
Reverse Transcriptases: From Discovery and Applications to Xenobiology.Chembiochem. 2023 Mar 1;24(5):e202200521. doi: 10.1002/cbic.202200521. Epub 2022 Nov 28. Chembiochem. 2023. PMID: 36354312 Review.
Cited by
-
Gene expression analysis of bone metastasis and circulating tumor cells from metastatic castrate-resistant prostate cancer patients.J Transl Med. 2016 Mar 15;14:72. doi: 10.1186/s12967-016-0829-5. J Transl Med. 2016. PMID: 26975354 Free PMC article.
-
Culture Conditions Affect Expression of DUX4 in FSHD Myoblasts.Molecules. 2015 May 8;20(5):8304-15. doi: 10.3390/molecules20058304. Molecules. 2015. PMID: 26007167 Free PMC article.
-
Gene Expression Profiling of FFPE Samples: A Titration Test.Technol Cancer Res Treat. 2022 Jan-Dec;21:15330338221129710. doi: 10.1177/15330338221129710. Technol Cancer Res Treat. 2022. PMID: 36415121 Free PMC article.
-
Use of chimeric influenza viruses as a novel internal control for diagnostic rRT-PCR assays.Appl Microbiol Biotechnol. 2016 Feb;100(4):1667-1676. doi: 10.1007/s00253-015-7042-y. Appl Microbiol Biotechnol. 2016. PMID: 26474983 Free PMC article.
-
Tutorial: Guidelines for Single-Cell RT-qPCR.Cells. 2021 Sep 30;10(10):2607. doi: 10.3390/cells10102607. Cells. 2021. PMID: 34685587 Free PMC article. Review.
References
-
- Taniguchi K, Kajiyama T, Kambara H. Quantitative analysis of gene expression in a single cell by qPCR. Nat Meth. 2009;6:503–506. - PubMed
-
- McKinney MD, Moon SJ, Kulesh DA, Larsen T, Schoepp RJ. Detection of viral RNA from paraffin-embedded tissues after prolonged formalin fixation. J Clin Virol. 2009;44:39–42. - PubMed
-
- Kawasaki E. Amplification of RNA. In: Innis MA, Gelfand DH, Sninsky JJ, While TJ, editors. PCR protocols: A guide to methods and applications. San Diego, CA: Academic Press; 1990.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

