The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover
- PMID: 21085673
- PMCID: PMC2978106
- DOI: 10.1371/journal.pone.0013935
The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover
Abstract
Background: Genetic evidence in Arabidopsis thaliana indicates that members of the Snf1-Related Kinases 2 family (SnRK2) are essential in mediating various stress-adaptive responses. Recent reports have indeed shown that one particular member, Open Stomata (OST)1, whose kinase activity is stimulated by the stress hormone abscisic acid (ABA), is a direct target of negative regulation by the core ABA co-receptor complex composed of PYR/PYL/RCAR and clade A Protein Phosphatase 2C (PP2C) proteins.
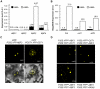
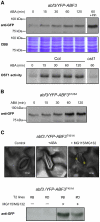
Methodology/principal findings: Here, the substrate preference of OST1 was interrogated at a genome-wide scale. We phosphorylated in vitro a bank of semi-degenerate peptides designed to assess the relative phosphorylation efficiency on a positionally fixed serine or threonine caused by systematic changes in the flanking amino acid sequence. Our results designate the ABA-responsive-element Binding Factor 3 (ABF3), which controls part of the ABA-regulated transcriptome, as a genuine OST1 substrate. Bimolecular Fluorescence Complementation experiments indicate that ABF3 interacts directly with OST1 in the nuclei of living plant cells. In vitro, OST1 phosphorylates ABF3 on multiple LXRXXpS/T preferred motifs including T451 located in the midst of a conserved 14-3-3 binding site. Using an antibody sensitive to the phosphorylated state of the preferred motif, we further show that ABF3 is phosphorylated on at least one such motif in response to ABA in vivo and that phospho-T451 is important for stabilization of ABF3.
Conclusions/significance: All together, our results suggest that OST1 phosphorylates ABF3 in vivo on T451 to create a 14-3-3 binding motif. In a wider physiological context, we propose that the long term responses to ABA that require sustained gene expression is, in part, mediated by the stabilization of ABFs driven by ABA-activated SnRK2s.
Conflict of interest statement
Figures




References
-
- Ton J, Flors V, Mauch-Mani B. The multifaceted role of ABA in disease resistance. Trends Plant Sci. 2009;14:310–317. - PubMed
-
- Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, et al. An update on abscisic acid signaling in plants and more. Molecular Plant. 2008;1:198–217. - PubMed
-
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

