Phospholipase D family member 4, a transmembrane glycoprotein with no phospholipase D activity, expression in spleen and early postnatal microglia
- PMID: 21085684
- PMCID: PMC2978679
- DOI: 10.1371/journal.pone.0013932
Phospholipase D family member 4, a transmembrane glycoprotein with no phospholipase D activity, expression in spleen and early postnatal microglia
Abstract
Background: Phospholipase D (PLD) catalyzes conversion of phosphatidylcholine into choline and phosphatidic acid, leading to a variety of intracellular signal transduction events. Two classical PLDs, PLD1 and PLD2, contain phosphatidylinositide-binding PX and PH domains and two conserved His-x-Lys-(x)(4)-Asp (HKD) motifs, which are critical for PLD activity. PLD4 officially belongs to the PLD family, because it possesses two HKD motifs. However, it lacks PX and PH domains and has a putative transmembrane domain instead. Nevertheless, little is known regarding expression, structure, and function of PLD4.
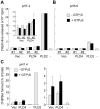
Methodology/principal findings: PLD4 was analyzed in terms of expression, structure, and function. Expression was analyzed in developing mouse brains and non-neuronal tissues using microarray, in situ hybridization, immunohistochemistry, and immunocytochemistry. Structure was evaluated using bioinformatics analysis of protein domains, biochemical analyses of transmembrane property, and enzymatic deglycosylation. PLD activity was examined by choline release and transphosphatidylation assays. Results demonstrated low to modest, but characteristic, PLD4 mRNA expression in a subset of cells preferentially localized around white matter regions, including the corpus callosum and cerebellar white matter, during the first postnatal week. These PLD4 mRNA-expressing cells were identified as Iba1-positive microglia. In non-neuronal tissues, PLD4 mRNA expression was widespread, but predominantly distributed in the spleen. Intense PLD4 expression was detected around the marginal zone of the splenic red pulp, and splenic PLD4 protein recovered from subcellular membrane fractions was highly N-glycosylated. PLD4 was heterologously expressed in cell lines and localized in the endoplasmic reticulum and Golgi apparatus. Moreover, heterologously expressed PLD4 proteins did not exhibit PLD enzymatic activity.
Conclusions/significance: Results showed that PLD4 is a non-PLD, HKD motif-carrying, transmembrane glycoprotein localized in the endoplasmic reticulum and Golgi apparatus. The spatiotemporally restricted expression patterns suggested that PLD4 might play a role in common function(s) among microglia during early postnatal brain development and splenic marginal zone cells.
Conflict of interest statement
Figures






References
-
- Exton JH. Regulation of phospholipase D. FEBS Lett. 2002;531:58–61. - PubMed
-
- Aoki J, Inoue A, Okudaira S. Two pathways for lysophosphatidic acid production. Biochim Biophys Acta. 2008;1781:513–518. - PubMed
-
- Roth MG. Molecular mechanisms of PLD function in membrane traffic. Traffic. 2008;9:1233–1239. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

