The roles of the Saccharomyces cerevisiae RecQ helicase SGS1 in meiotic genome surveillance
- PMID: 21085703
- PMCID: PMC2976770
- DOI: 10.1371/journal.pone.0015380
The roles of the Saccharomyces cerevisiae RecQ helicase SGS1 in meiotic genome surveillance
Abstract
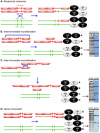
Background: The Saccharomyces cerevisiae RecQ helicase Sgs1 is essential for mitotic and meiotic genome stability. The stage at which Sgs1 acts during meiosis is subject to debate. Cytological experiments showed that a deletion of SGS1 leads to an increase in synapsis initiation complexes and axial associations leading to the proposal that it has an early role in unwinding surplus strand invasion events. Physical studies of recombination intermediates implicate it in the dissolution of double Holliday junctions between sister chromatids.
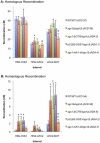
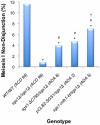
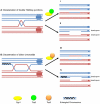
Methodology/principal findings: In this work, we observed an increase in meiotic recombination between diverged sequences (homeologous recombination) and an increase in unequal sister chromatid events when SGS1 is deleted. The first of these observations is most consistent with an early role of Sgs1 in unwinding inappropriate strand invasion events while the second is consistent with unwinding or dissolution of recombination intermediates in an Mlh1- and Top3-dependent manner. We also provide data that suggest that Sgs1 is involved in the rejection of 'second strand capture' when sequence divergence is present. Finally, we have identified a novel class of tetrads where non-sister spores (pairs of spores where each contains a centromere marker from a different parent) are inviable. We propose a model for this unusual pattern of viability based on the inability of sgs1 mutants to untangle intertwined chromosomes. Our data suggest that this role of Sgs1 is not dependent on its interaction with Top3. We propose that in the absence of SGS1 chromosomes may sometimes remain entangled at the end of pre-meiotic replication. This, combined with reciprocal crossing over, could lead to physical destruction of the recombined and entangled chromosomes. We hypothesise that Sgs1, acting in concert with the topoisomerase Top2, resolves these structures.
Conclusions: This work provides evidence that Sgs1 interacts with various partner proteins to maintain genome stability throughout meiosis.
Conflict of interest statement
Figures


References
-
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. - PubMed
-
- Sun H, Treco D, Szostak JW. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991;64:1155–1161. - PubMed
-
- Shinohara A, Shinohara M. Roles of RecA homologues Rad51 and Dmc1 during meiotic recombination. Cytogenet Genome Res. 2004;107:201–207. - PubMed
-
- Hunter N, Kleckner N. The Single-End Invasion: An Asymmetric Intermediate at the Double-Strand Break to Double-Holliday Junction Transition of Meiotic Recombination. Cell. 2001;106:59–70. - PubMed
-
- Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

