Topoisomerases and site-specific recombinases: similarities in structure and mechanism
- PMID: 21087076
- PMCID: PMC6290911
- DOI: 10.3109/10409238.2010.513375
Topoisomerases and site-specific recombinases: similarities in structure and mechanism
Abstract
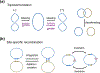
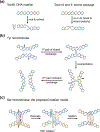
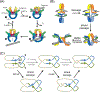
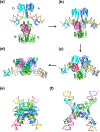
The processes of DNA topoisomerization and site-specific recombination are fundamentally similar: DNA cleavage by forming a phospho-protein covalent linkage, DNA topological rearrangement, and DNA ligation coupled with protein regeneration. Type IB DNA topoisomerases are structurally and mechanistically homologous to tyrosine recombinases. Both enzymes nick DNA double helices independent of metal ions, form 3'-phosphotyrosine intermediates, and rearrange the free 5' ends relative to the uncut strands by swiveling. In contrast, serine recombinases generate 5'-phospho-serine intermediates. A 180° relative rotation of the two halves of a 100 kDa terameric serine recombinase and DNA complex has been proposed as the mechanism of strand exchange. Here I propose an alternative mechanism. Interestingly, the catalytic domain of serine recombinases has structural similarity to the TOPRIM domain, conserved among all Type IA and Type II topoisomerases and responsible for metal binding and DNA cleavage. TOPRIM topoisomerases also cleave DNA to generate 5'-phosphate and 3'-OH groups. Based on the existing biochemical data and crystal structures of topoisomerase II and serine recombinases bound to pre- and post-cleavage DNA, I suggest a strand passage mechanism for DNA recombination by serine recombinases. This mechanism is reminiscent of DNA topoisomerization and does not require subunit rotation.
Conflict of interest statement
Declaration of Interest
The research was funded by the intramural research program of NIDDK, NIH.
Declaration of no interest
The author reports no conflicts of interest. The author alone is responsible for the content and writing of the paper.
Figures


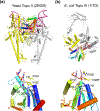
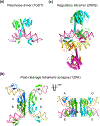
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
