Classification of rice (Oryza sativa L. Japonica nipponbare) immunophilins (FKBPs, CYPs) and expression patterns under water stress
- PMID: 21087465
- PMCID: PMC3012604
- DOI: 10.1186/1471-2229-10-253
Classification of rice (Oryza sativa L. Japonica nipponbare) immunophilins (FKBPs, CYPs) and expression patterns under water stress
Abstract
Background: FK506 binding proteins (FKBPs) and cyclophilins (CYPs) are abundant and ubiquitous proteins belonging to the peptidyl-prolyl cis/trans isomerase (PPIase) superfamily, which regulate much of metabolism through a chaperone or an isomerization of proline residues during protein folding. They are collectively referred to as immunophilin (IMM), being present in almost all cellular organs. In particular, a number of IMMs relate to environmental stresses.
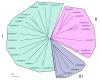
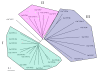
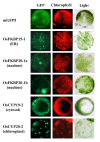
Results: FKBP and CYP proteins in rice (Oryza sativa cv. Japonica) were identified and classified, and given the appropriate name for each IMM, considering the ortholog-relation with Arabidopsis and Chlamydomonas or molecular weight of the proteins. 29 FKBP and 27 CYP genes can putatively be identified in rice; among them, a number of genes can be putatively classified as orthologs of Arabidopsis IMMs. However, some genes were novel, did not match with those of Arabidopsis and Chlamydomonas, and several genes were paralogs by genetic duplication. Among 56 IMMs in rice, a significant number are regulated by salt and/or desiccation stress. In addition, their expression levels responding to the water-stress have been analyzed in different tissues, and some subcellular IMMs located by means of tagging with GFP protein.
Conclusion: Like other green photosynthetic organisms such as Arabidopsis (23 FKBPs and 29 CYPs) and Chlamydomonas (23 FKBs and 26 CYNs), rice has the highest number of IMM genes among organisms reported so far, suggesting that the numbers relate closely to photosynthesis. Classification of the putative FKBPs and CYPs in rice provides the information about their evolutional/functional significance when comparisons are drawn with the relatively well studied genera, Arabidopsis and Chlamydomonas. In addition, many of the genes upregulated by water stress offer the possibility of manipulating the stress responses in rice.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

