Comparative study of unsupervised dimension reduction techniques for the visualization of microarray gene expression data
- PMID: 21087509
- PMCID: PMC2998530
- DOI: 10.1186/1471-2105-11-567
Comparative study of unsupervised dimension reduction techniques for the visualization of microarray gene expression data
Abstract
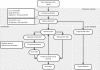
Background: Visualization of DNA microarray data in two or three dimensional spaces is an important exploratory analysis step in order to detect quality issues or to generate new hypotheses. Principal Component Analysis (PCA) is a widely used linear method to define the mapping between the high-dimensional data and its low-dimensional representation. During the last decade, many new nonlinear methods for dimension reduction have been proposed, but it is still unclear how well these methods capture the underlying structure of microarray gene expression data. In this study, we assessed the performance of the PCA approach and of six nonlinear dimension reduction methods, namely Kernel PCA, Locally Linear Embedding, Isomap, Diffusion Maps, Laplacian Eigenmaps and Maximum Variance Unfolding, in terms of visualization of microarray data.
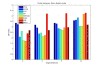
Results: A systematic benchmark, consisting of Support Vector Machine classification, cluster validation and noise evaluations was applied to ten microarray and several simulated datasets. Significant differences between PCA and most of the nonlinear methods were observed in two and three dimensional target spaces. With an increasing number of dimensions and an increasing number of differentially expressed genes, all methods showed similar performance. PCA and Diffusion Maps responded less sensitive to noise than the other nonlinear methods.
Conclusions: Locally Linear Embedding and Isomap showed a superior performance on all datasets. In very low-dimensional representations and with few differentially expressed genes, these two methods preserve more of the underlying structure of the data than PCA, and thus are favorable alternatives for the visualization of microarray data.
Figures




References
-
- Lim IS, Ciechomski PDH, Sarni S, Thalmann D. Planar arrangement of high-dimensional biomedical data sets by Isomap coordinates. In Proceedings of the 16 th IEEE Symposium on Computer-Based Medical Systems. 2003. pp. 50–55.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

