Insights into the mechanism of type I dehydroquinate dehydratases from structures of reaction intermediates
- PMID: 21087925
- PMCID: PMC3030358
- DOI: 10.1074/jbc.M110.192831
Insights into the mechanism of type I dehydroquinate dehydratases from structures of reaction intermediates
Erratum in
-
Insights into the mechanism of type I dehydroquinate dehydratases from structures of reaction intermediates.J Biol Chem. 2015 Jul 31;290(31):19008. doi: 10.1074/jbc.A110.192831. J Biol Chem. 2015. PMID: 26232400 Free PMC article. No abstract available.
Abstract
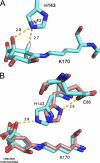
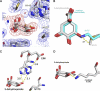
The biosynthetic shikimate pathway consists of seven enzymes that catalyze sequential reactions to generate chorismate, a critical branch point in the synthesis of the aromatic amino acids. The third enzyme in the pathway, dehydroquinate dehydratase (DHQD), catalyzes the dehydration of 3-dehydroquinate to 3-dehydroshikimate. We present three crystal structures of the type I DHQD from the intestinal pathogens Clostridium difficile and Salmonella enterica. Structures of the enzyme with substrate and covalent pre- and post-dehydration reaction intermediates provide snapshots of successive steps along the type I DHQD-catalyzed reaction coordinate. These structures reveal that the position of the substrate within the active site does not appreciably change upon Schiff base formation. The intermediate state structures reveal a reaction state-dependent behavior of His-143 in which the residue adopts a conformation proximal to the site of catalytic dehydration only when the leaving group is present. We speculate that His-143 is likely to assume differing catalytic roles in each of its observed conformations. One conformation of His-143 positions the residue for the formation/hydrolysis of the covalent Schiff base intermediates, whereas the other conformation positions the residue for a role in the catalytic dehydration event. The fact that the shikimate pathway is absent from humans makes the enzymes of the pathway potential targets for the development of non-toxic antimicrobials. The structures and mechanistic insight presented here may inform the design of type I DHQD enzyme inhibitors.
Figures
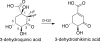


Similar articles
-
A conserved surface loop in type I dehydroquinate dehydratases positions an active site arginine and functions in substrate binding.Biochemistry. 2011 Mar 29;50(12):2357-63. doi: 10.1021/bi102020s. Epub 2011 Feb 21. Biochemistry. 2011. PMID: 21291284 Free PMC article.
-
Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of Schiff base formation.Biochemistry. 2014 Feb 11;53(5):872-80. doi: 10.1021/bi4015506. Epub 2014 Jan 31. Biochemistry. 2014. PMID: 24437575 Free PMC article.
-
New insights into the mechanism of the Schiff base hydrolysis catalyzed by type I dehydroquinate dehydratase from S. enterica: a theoretical study.Org Biomol Chem. 2012 Sep 21;10(35):7037-44. doi: 10.1039/c2ob25605c. Epub 2012 Jul 31. Org Biomol Chem. 2012. PMID: 22847490
-
The pre-chorismate (shikimate) and quinate pathways in filamentous fungi: theoretical and practical aspects.J Gen Microbiol. 1993 Dec;139(12):2891-9. doi: 10.1099/00221287-139-12-2891. J Gen Microbiol. 1993. PMID: 8126417 Review. No abstract available.
-
The shikimate pathway: review of amino acid sequence, function and three-dimensional structures of the enzymes.Crit Rev Microbiol. 2015 Jun;41(2):172-89. doi: 10.3109/1040841X.2013.813901. Epub 2013 Aug 6. Crit Rev Microbiol. 2015. PMID: 23919299 Review.
Cited by
-
Reassessing the type I dehydroquinate dehydratase catalytic triad: kinetic and structural studies of Glu86 mutants.Protein Sci. 2013 Apr;22(4):418-24. doi: 10.1002/pro.2218. Epub 2013 Feb 11. Protein Sci. 2013. PMID: 23341204 Free PMC article.
-
Adherence to Bürgi-Dunitz stereochemical principles requires significant structural rearrangements in Schiff-base formation: insights from transaldolase complexes.Acta Crystallogr D Biol Crystallogr. 2014 Feb;70(Pt 2):544-52. doi: 10.1107/S1399004713030666. Epub 2014 Jan 31. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 24531488 Free PMC article.
-
A conserved surface loop in type I dehydroquinate dehydratases positions an active site arginine and functions in substrate binding.Biochemistry. 2011 Mar 29;50(12):2357-63. doi: 10.1021/bi102020s. Epub 2011 Feb 21. Biochemistry. 2011. PMID: 21291284 Free PMC article.
-
Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.Life Sci Alliance. 2022 May 5;5(8):e202101358. doi: 10.26508/lsa.202101358. Print 2022 Aug. Life Sci Alliance. 2022. PMID: 35512834 Free PMC article.
-
Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of Schiff base formation.Biochemistry. 2014 Feb 11;53(5):872-80. doi: 10.1021/bi4015506. Epub 2014 Jan 31. Biochemistry. 2014. PMID: 24437575 Free PMC article.
References
-
- Bentley R. (1990) Crit. Rev. Biochem. Mol. Biol. 25, 307–384 - PubMed
-
- Kishore G. M., Shah D. M. (1988) Annu. Rev. Biochem. 57, 627–663 - PubMed
-
- Marques M. R., Pereira J. H., Oliveira J. S., Basso L. A., de Azevedo W. F., Jr., Santos D. S., Palma M. S. (2007) Curr. Drug Targets 8, 445–457 - PubMed
-
- Noble M., Sinha Y., Kolupaev A., Demin O., Earnshaw D., Tobin F., West J., Martin J. D., Qiu C., Liu W. S., DeWolf W. E., Jr., Tew D., Goryanin I. I. (2006) Biotechnol. Bioeng. 95, 560–573 - PubMed
-
- Butler J. R., Alworth W. L., Nugent M. J. (1974) J. Am. Chem. Soc. 96, 1617–1618
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

