Role of RecJ-like protein with 5'-3' exonuclease activity in oligo(deoxy)nucleotide degradation
- PMID: 21087930
- PMCID: PMC3024776
- DOI: 10.1074/jbc.M110.161596
Role of RecJ-like protein with 5'-3' exonuclease activity in oligo(deoxy)nucleotide degradation
Abstract
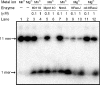
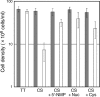
RecJ-like proteins belonging to the DHH family have been proposed to function as oligoribonucleases and 3'-phosphoadenosine 5'-phosphate (pAp) phosphatases in bacteria and archaea, which do not have Orn (oligoribonuclease) and CysQ (pAp phosphatase) homologs. In this study, we analyzed the biochemical and physiological characterization of the RecJ-like protein TTHA0118 from Thermus thermophilus HB8. TTHA0118 had high enzymatic activity as an oligodeoxyribonucleotide- and oligoribonucleotide-specific exonuclease and as pAp phosphatase. The polarity of degradation was 5' to 3', in contrast to previous reports about Bacillus subtilis NrnA, a RecJ-like protein. TTHA0118 preferentially hydrolyzed short oligodeoxyribonucleotides and oligoribonucleotides, whereas the RecJ exonuclease from T. thermophilus HB8 showed no such length dependence on oligodeoxyribonucleotide substrates. An insertion mutation of the ttha0118 gene led to growth reduction in minimum essential medium. Added 5'-mononucleotides, nucleosides, and cysteine increased growth of the ttha0118 mutant in minimum essential medium. The RecJ-like protein Mpn140 from Mycoplasma pneumoniae M129, which cannot synthesize nucleic acid precursors de novo, showed similar biochemical features to TTHA0118. Furthermore, B. subtilis NrnA also hydrolyzed oligo(deoxy)ribonucleotides in a 5'-3' direction. These results suggested that these RecJ-like proteins act in recycling short oligonucleotides to mononucleotides and in controlling pAp concentrations in vivo.
Figures





Similar articles
-
Structure of RecJ exonuclease defines its specificity for single-stranded DNA.J Biol Chem. 2010 Mar 26;285(13):9762-9769. doi: 10.1074/jbc.M109.096487. Epub 2010 Feb 2. J Biol Chem. 2010. PMID: 20129927 Free PMC article.
-
A novel single-stranded DNA-specific 3'-5' exonuclease, Thermus thermophilus exonuclease I, is involved in several DNA repair pathways.Nucleic Acids Res. 2010 Sep;38(17):5692-705. doi: 10.1093/nar/gkq350. Epub 2010 May 10. Nucleic Acids Res. 2010. PMID: 20457749 Free PMC article.
-
Overexpression, purification and characterization of RecJ protein from Thermus thermophilus HB8 and its core domain.Nucleic Acids Res. 2001 Nov 15;29(22):4617-24. doi: 10.1093/nar/29.22.4617. Nucleic Acids Res. 2001. PMID: 11713311 Free PMC article.
-
Biochemical characterisation of UvrD helicase and RecJ exonuclease from Neisseria gonorrhoeae.Int J Biol Macromol. 2025 May;306(Pt 4):141530. doi: 10.1016/j.ijbiomac.2025.141530. Epub 2025 Mar 1. Int J Biol Macromol. 2025. PMID: 40032130
-
Nano-RNases: oligo- or dinucleases?FEMS Microbiol Rev. 2022 Nov 2;46(6):fuac038. doi: 10.1093/femsre/fuac038. FEMS Microbiol Rev. 2022. PMID: 36026528 Free PMC article. Review.
Cited by
-
Functional Analysis of a c-di-AMP-specific Phosphodiesterase MsPDE from Mycobacterium smegmatis.Int J Biol Sci. 2015 May 30;11(7):813-24. doi: 10.7150/ijbs.11797. eCollection 2015. Int J Biol Sci. 2015. PMID: 26078723 Free PMC article.
-
The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.Nucleic Acids Res. 2017 Dec 1;45(21):12551-12564. doi: 10.1093/nar/gkx887. Nucleic Acids Res. 2017. PMID: 30053256 Free PMC article.
-
Characterization of the MSMEG_2631 gene (mmp) encoding a multidrug and toxic compound extrusion (MATE) family protein in Mycobacterium smegmatis and exploration of its polyspecific nature using biolog phenotype microarray.J Bacteriol. 2013 Apr;195(7):1610-21. doi: 10.1128/JB.01724-12. Epub 2013 Jan 4. J Bacteriol. 2013. PMID: 23292779 Free PMC article.
-
Crystal structure of the ligand-binding form of nanoRNase from Bacteroides fragilis, a member of the DHH/DHHA1 phosphoesterase family of proteins.FEBS Lett. 2013 Aug 19;587(16):2669-74. doi: 10.1016/j.febslet.2013.06.053. Epub 2013 Jul 9. FEBS Lett. 2013. PMID: 23851074 Free PMC article.
-
Characterization of NrnA homologs from Mycobacterium tuberculosis and Mycoplasma pneumoniae.RNA. 2012 Jan;18(1):155-65. doi: 10.1261/rna.029132.111. Epub 2011 Nov 23. RNA. 2012. PMID: 22114320 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

