Chemoreceptors in signalling complexes: shifted conformation and asymmetric coupling
- PMID: 21091513
- PMCID: PMC3010867
- DOI: 10.1111/j.1365-2958.2010.07408.x
Chemoreceptors in signalling complexes: shifted conformation and asymmetric coupling
Abstract
Bacterial chemotaxis is mediated by signalling complexes of chemoreceptors, histidine kinase CheA and coupling protein CheW. Interactions in complexes profoundly affect the kinase. We investigated effects of these interactions on chemoreceptors by comparing receptors alone and in complexes. Assays of initial rates of methylation indicated that signalling complexes shifted receptor conformation towards the methylation-on, higher-ligand-affinity, kinase-off state, tuning receptors for greater sensitivity. In contrast, transmembrane and conformational signalling within chemoreceptors was essentially unaltered, consistent with other evidence identifying receptor dimers as the fundamental units of such signalling. In signalling complexes, coupling of ligand binding to kinase activity is cooperative and the dynamic range of kinase control expanded > 100-fold by receptor adaptational modification. We observed no cooperativity in influence of ligand on receptor conformation, only on kinase activity. However, receptor modification generated increased dynamic range in a stepwise fashion, partly in coupling ligand to receptor conformation and partly in coupling receptor conformation to kinase activity. Thus, receptors and kinase were not equivalently affected by interactions in signalling complexes or by ligand binding and adaptational modification, indicating asymmetrical coupling between them. This has implications for mechanisms of precise adaptation. Coupling might vary, providing a previously unappreciated locus for sensory control.
© 2010 Blackwell Publishing Ltd.
Figures
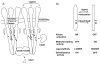


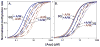


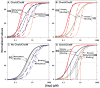
References
-
- Alon U, Surette MG, Barkai N, Leibler S. Robustness in Bacterial Chemotaxis. Nature. 1999;397:168–171. - PubMed
-
- Bhatnagar J, Borbat PP, Pollard AM, Bilwes AM, Freed JH, Crane BR. Structure of the Ternary Complex Formed by a Chemotaxis Receptor Signaling Domain, the CheA Histidine Kinase, and the Coupling Protein CheW As Determined by Pulsed Dipolar ESR Spectroscopy. Biochemistry. 2010;49:3824–3841. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

