Imprinted genes show unique patterns of sequence conservation
- PMID: 21092170
- PMCID: PMC3091771
- DOI: 10.1186/1471-2164-11-649
Imprinted genes show unique patterns of sequence conservation
Abstract
Background: Genomic imprinting is an evolutionary conserved mechanism of epigenetic gene regulation in placental mammals that results in silencing of one of the parental alleles. In order to decipher interactions between allele-specific DNA methylation of imprinted genes and evolutionary conservation, we performed a genome-wide comparative investigation of genomic sequences and highly conserved elements of imprinted genes in human and mouse.
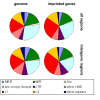
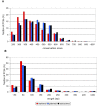
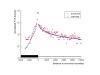
Results: Evolutionarily conserved elements in imprinted regions differ from those associated with autosomal genes in various ways. Whereas for maternally expressed genes strong divergence of protein-encoding sequences is most prominent, paternally expressed genes exhibit substantial conservation of coding and noncoding sequences. Conserved elements in imprinted regions are marked by enrichment of CpG dinucleotides and low (TpG+CpA)/(2·CpG) ratios indicate reduced CpG deamination. Interestingly, paternally and maternally expressed genes can be distinguished by differences in G+C and CpG contents that might be associated with unusual epigenetic features. Especially noncoding conserved elements of paternally expressed genes are exceptionally G+C and CpG rich. In addition, we confirmed a frequent occurrence of intronic CpG islands and observed a decelerated degeneration of ancient LINE-1 repeats. We also found a moderate enrichment of YY1 and CTCF binding sites in imprinted regions and identified several short sequence motifs in highly conserved elements that might act as additional regulatory elements.
Conclusions: We discovered several novel conserved DNA features that might be related to allele-specific DNA methylation. Our results hint at reduced CpG deamination rates in imprinted regions, which affects mostly noncoding conserved elements of paternally expressed genes. Pronounced differences between maternally and paternally expressed genes imply specific modes of evolution as a result of differences in epigenetic features and a special response to selective pressure. In addition, our data support the potential role of intronic CpG islands as epigenetic key regulatory elements and suggest that evolutionary conserved LINE-1 elements fulfill regulatory functions in imprinted regions.
Figures
Similar articles
-
Tandem repeats in the CpG islands of imprinted genes.Genomics. 2006 Sep;88(3):323-32. doi: 10.1016/j.ygeno.2006.03.019. Epub 2006 May 11. Genomics. 2006. PMID: 16690248
-
A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes.Genome Res. 2002 Apr;12(4):543-54. doi: 10.1101/gr.224102. Genome Res. 2002. PMID: 11932239 Free PMC article.
-
Epigenetic analysis of the Dlk1-Gtl2 imprinted domain on mouse chromosome 12: implications for imprinting control from comparison with Igf2-H19.Hum Mol Genet. 2002 Jan 1;11(1):77-86. doi: 10.1093/hmg/11.1.77. Hum Mol Genet. 2002. PMID: 11773001
-
Imprinting of the mouse Igf2r gene depends on an intronic CpG island.Mol Cell Endocrinol. 1998 May 25;140(1-2):9-14. doi: 10.1016/s0303-7207(98)00022-7. Mol Cell Endocrinol. 1998. PMID: 9722161 Review.
-
Imprinted genes and human disease: an evolutionary perspective.Adv Exp Med Biol. 2008;626:101-15. Adv Exp Med Biol. 2008. PMID: 18372794 Review.
Cited by
-
Importance of the matriline for genomic imprinting, brain development and behaviour.Philos Trans R Soc Lond B Biol Sci. 2013 Jan 5;368(1609):20110327. doi: 10.1098/rstb.2011.0327. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23166391 Free PMC article. Review.
-
Conservation of Repeats at the Mammalian KCNQ1OT1-CDKN1C Region Suggests a Role in Genomic Imprinting.Evol Bioinform Online. 2017 Jun 16;13:1176934317715238. doi: 10.1177/1176934317715238. eCollection 2017. Evol Bioinform Online. 2017. PMID: 28659711 Free PMC article.
-
Cellular functions of genetically imprinted genes in human and mouse as annotated in the gene ontology.PLoS One. 2012;7(11):e50285. doi: 10.1371/journal.pone.0050285. Epub 2012 Nov 30. PLoS One. 2012. PMID: 23226257 Free PMC article.
-
Epigenetic modifications potentially controlling the allelic expression of imprinted genes in sunflower endosperm.BMC Plant Biol. 2021 Dec 4;21(1):570. doi: 10.1186/s12870-021-03344-4. BMC Plant Biol. 2021. PMID: 34863098 Free PMC article.
-
The Role of Long Non-coding RNAs in Human Imprinting Disorders: Prospective Therapeutic Targets.Front Cell Dev Biol. 2021 Oct 25;9:730014. doi: 10.3389/fcell.2021.730014. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34760887 Free PMC article. Review.
References
-
- Kobayashi H, Suda C, Abe T, Kohara Y, Ikemura T, Sasaki H. Bisulfite sequencing and dinucleotide content analysis of 15 imprinted mouse differentially methylated regions (DMRs): paternally methylated DMRs contain less CpGs than maternally methylated DMRs. Cytogenet Genome Res. 2006;113:130–137. doi: 10.1159/000090824. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

