Identification and characterization of a novel fumarase gene by metagenome expression cloning from marine microorganisms
- PMID: 21092234
- PMCID: PMC3002918
- DOI: 10.1186/1475-2859-9-91
Identification and characterization of a novel fumarase gene by metagenome expression cloning from marine microorganisms
Abstract
Background: Fumarase catalyzes the reversible hydration of fumarate to L-malate and is a key enzyme in the tricarboxylic acid (TCA) cycle and in amino acid metabolism. Fumarase is also used for the industrial production of L-malate from the substrate fumarate. Thermostable and high-activity fumarases from organisms that inhabit extreme environments may have great potential in industry, biotechnology, and basic research. The marine environment is highly complex and considered one of the main reservoirs of microbial diversity on the planet. However, most of the microorganisms are inaccessible in nature and are not easily cultivated in the laboratory. Metagenomic approaches provide a powerful tool to isolate and identify enzymes with novel biocatalytic activities for various biotechnological applications.
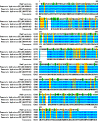
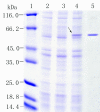
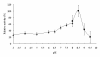
Results: A plasmid metagenomic library was constructed from uncultivated marine microorganisms within marine water samples. Through sequence-based screening of the DNA library, a gene encoding a novel fumarase (named FumF) was isolated. Amino acid sequence analysis revealed that the FumF protein shared the greatest homology with Class II fumarate hydratases from Bacteroides sp. 2_1_33B and Parabacteroides distasonis ATCC 8503 (26% identical and 43% similar). The putative fumarase gene was subcloned into pETBlue-2 vector and expressed in E. coli BL21(DE3)pLysS. The recombinant protein was purified to homogeneity. Functional characterization by high performance liquid chromatography confirmed that the recombinant FumF protein catalyzed the hydration of fumarate to form L-malate. The maximum activity for FumF protein occurred at pH 8.5 and 55°C in 5 mM Mg(2+). The enzyme showed higher affinity and catalytic efficiency under optimal reaction conditions: K(m) = 0.48 mM, V(max) = 827 μM/min/mg, and k(cat)/K(m) = 1900 mM/s.
Conclusions: We isolated a novel fumarase gene, fumF, from a sequence-based screen of a plasmid metagenomic library from uncultivated marine microorganisms. The properties of FumF protein may be ideal for the industrial production of L-malate under higher temperature conditions. The identification of FumF underscores the potential of marine metagenome screening for novel biomolecules.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

