Epigenetic control of the basal-like gene expression profile via Interleukin-6 in breast cancer cells
- PMID: 21092249
- PMCID: PMC3002335
- DOI: 10.1186/1476-4598-9-300
Epigenetic control of the basal-like gene expression profile via Interleukin-6 in breast cancer cells
Abstract
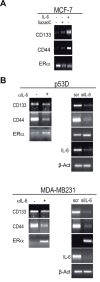
Background: Basal-like carcinoma are aggressive breast cancers that frequently carry p53 inactivating mutations, lack estrogen receptor-α (ERα) and express the cancer stem cell markers CD133 and CD44. These tumors also over-express Interleukin 6 (IL-6), a pro-inflammatory cytokine that stimulates the growth of breast cancer stem/progenitor cells.
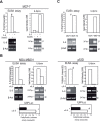
Results: Here we show that p53 deficiency in breast cancer cells induces a loss of methylation at IL-6 proximal promoter region, which is maintained by an IL-6 autocrine loop. IL-6 also elicits the loss of methylation at the CD133 promoter region 1 and of CD44 proximal promoter, enhancing CD133 and CD44 gene transcription. In parallel, IL-6 induces the methylation of estrogen receptor (ERα) promoter and the loss of ERα mRNA expression. Finally, IL-6 induces the methylation of IL-6 distal promoter and of CD133 promoter region 2, which harbour putative repressor regions.
Conclusion: We conclude that IL-6, whose methylation-dependent autocrine loop is triggered by the inactivation of p53, induces an epigenetic reprogramming that drives breast carcinoma cells towards a basal-like/stem cell-like gene expression profile.
Figures






References
-
- Bertheau P, Turpin E, Rickman DS, Espié M, de Reyniès A, Feugeas Plassa LF, Soliman H, Varna M, de Roquancourt A, Lehmann-Che J, Beuzard Y, Marty M, Misset JL, Janin A, de Thé H. Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen. PLOS MED. 2007;4:585–593. doi: 10.1371/journal.pmed.0040090. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

