Independent evolution of the core and accessory gene sets in the genus Neisseria: insights gained from the genome of Neisseria lactamica isolate 020-06
- PMID: 21092259
- PMCID: PMC3091772
- DOI: 10.1186/1471-2164-11-652
Independent evolution of the core and accessory gene sets in the genus Neisseria: insights gained from the genome of Neisseria lactamica isolate 020-06
Abstract
Background: The genus Neisseria contains two important yet very different pathogens, N. meningitidis and N. gonorrhoeae, in addition to non-pathogenic species, of which N. lactamica is the best characterized. Genomic comparisons of these three bacteria will provide insights into the mechanisms and evolution of pathogenesis in this group of organisms, which are applicable to understanding these processes more generally.
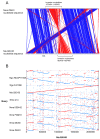
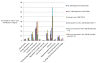
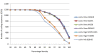
Results: Non-pathogenic N. lactamica exhibits very similar population structure and levels of diversity to the meningococcus, whilst gonococci are essentially recent descendents of a single clone. All three species share a common core gene set estimated to comprise around 1190 CDSs, corresponding to about 60% of the genome. However, some of the nucleotide sequence diversity within this core genome is particular to each group, indicating that cross-species recombination is rare in this shared core gene set. Other than the meningococcal cps region, which encodes the polysaccharide capsule, relatively few members of the large accessory gene pool are exclusive to one species group, and cross-species recombination within this accessory genome is frequent.
Conclusion: The three Neisseria species groups represent coherent biological and genetic groupings which appear to be maintained by low rates of inter-species horizontal genetic exchange within the core genome. There is extensive evidence for exchange among positively selected genes and the accessory genome and some evidence of hitch-hiking of housekeeping genes with other loci. It is not possible to define a 'pathogenome' for this group of organisms and the disease causing phenotypes are therefore likely to be complex, polygenic, and different among the various disease-associated phenotypes observed.
Figures

References
-
- Morse SA, Knapp JS. In: The Prokaryotes. 2. Balows A, Trüper HG, Dworkin M, Harder W, Schleifer K-H, editor. New York: Springer-Verlag; 1992. The Genus Neisseria; pp. 2495–2559.
-
- Perrin A, Bonacorsi S, Carbonnelle E, Talibi D, Dessen P, Nassif X, Tinsley C. Comparative genomics identifies the genetic islands that distinguish Neisseria meningitidis, the agent of cerebrospinal meningitis, from other Neisseria species. Infect Immun. 2002;70(12):7063–7072. doi: 10.1128/IAI.70.12.7063-7072.2002. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

